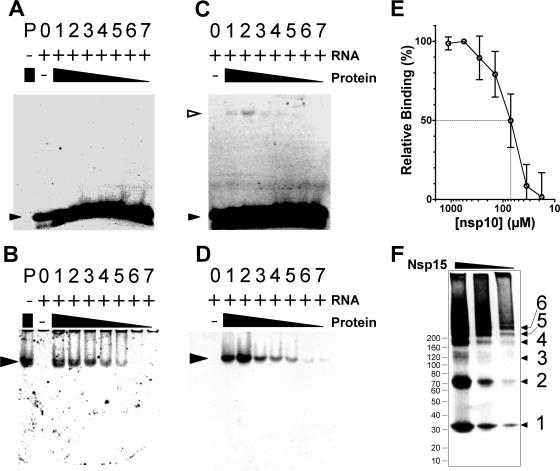
FIG. 1.
Electrophoretic mobility shift assays. Monomeric SARS-CoV nsp15 (A and B) and native nsp10 (C and D) were incubated with fluoresceinated ssRNA substrate, and complexes were separated by native electrophoresis on 6% polyacrylamide gels. Fluoresceinated RNA was visualized under UV illumination (A and C), and protein was visualized similarly after SYPRO-ruby poststaining (B and D). Lanes: P, protein only; 0, RNA only; 1 to 7, gradient of protein with a fixed amount of RNA. Free RNA is indicated with a small filled triangle, protein is indicated with a large filled triangle, and protein-RNA complexes are indicated with open triangles. (E) nsp10 was used as a positive control; its binding affinity to the fluoresceinated RNA substrate is shown. The dotted lines indicate the concentration of nsp10 that correlates to 50% binding. (F) PFO-PAGE gel indicating that trimmed nsp15 exists primarily as a monomer and as a dimer; although a variety of larger forms exists, this construct appears to be specifically defective in trimer formation.