Abstract
Previous studies of Epstein-Barr virus (EBV) replication focused mainly on the viral and cellular factors involved in replication compartment assembly and controlling the cell cycle. However, little is known about how EBV reorganizes nuclear architecture and the chromatin territories. In EBV-positive nasopharyngeal carcinoma NA cells or Akata cells, we noticed that cellular chromatin becomes highly condensed upon EBV reactivation. In searching for the possible mechanisms involved, we found that transient expression of EBV BGLF4 kinase induces unscheduled chromosome condensation, nuclear lamina disassembly, and stress fiber rearrangements, independently of cellular DNA replication and Cdc2 activity. BGLF4 interacts with condensin complexes, the major components in mitotic chromosome assembly, and induces condensin phosphorylation at Cdc2 consensus motifs. BGLF4 also stimulates the decatenation activity of topoisomerase II, suggesting that it may induce chromosome condensation through condensin and topoisomerase II activation. The ability to induce chromosome condensation is conserved in another gammaherpesvirus kinase, murine herpesvirus 68 ORF36. Together, these findings suggest a novel mechanism by which gammaherpesvirus kinases may induce multiple premature mitotic events to provide more extrachromosomal space for viral DNA replication and successful egress of nucleocapsid from the nucleus.
DNA viruses adopt various strategies to facilitate their replication and maturation within host cells, including usurping the cellular DNA replication machinery and taking over the nuclear space for viral DNA replication, transcription, and packaging. Small DNA viruses, such as simian virus 40 and papillomaviruses, modulate the cell cycle control pathway and promote entry into S phase. This enables the host DNA polymerase and the increased nucleotide pool to be used for virus replication. For large DNA viruses, such as herpesviruses, which encode DNA replication and nucleotide metabolism enzymes, the viral DNA replication strategy is controlled through even more sophisticated interactions between host and viral machineries (reviewed in references 36 and 57).
Epstein-Barr virus (EBV) belongs to the Gammaherpesvirinae and infects most of the human population worldwide. Infection may cause infectious mononucleosis and is closely associated with human malignant diseases, such as nasopharyngeal carcinoma (NPC) and Burkitt's lymphoma (61). Two different mechanisms have evolved to sustain successful infection of EBV. After primary infection, EBV becomes latent and the virally encoded EBNA-1 ensures that the circular episomal genome replicates during the S phase of the cell cycle and partitions equally into the daughter cells at mitosis (38). Upon immunoglobulin (Ig) cross-linking or chemical stimulation, EBV can be reactivated and express two immediate-early transactivators, Zta and Rta, which then turn on a cascade of viral gene expression to initiate lytic virus replication. Simultaneously, Zta and Rta may also modulate the cell cycle to facilitate virus replication (41). Recombinant adeno-Rta is able to induce expression of E2F1 and decrease expression of Rb, p107, and p130, suggesting that some S-phase factors are required for viral DNA replication (54). Moreover, S-phase cyclin-dependent kinase activities increase during virus replication, while cellular chromosomal DNA replication is inhibited (30, 31). Phosphorylation of the cellular replication origin binding complex MCM4-MCM6-MCM7 during EBV lytic replication thus inactivates the MCM4 helicase activity to prevent the initiation of cellular DNA replication (29).
In addition to hijacking the resources for cellular DNA replication, the virus needs to take over the compact chromatin territories in the nucleus for robust lytic viral DNA replication. In the case of the well-studied alphaherpesvirus herpes simplex virus type 1 (HSV-1), the viral replication compartments gradually become large globular structures, coupled with compression and marginalization of cellular chromatin (42). Finally, to complete virus encapsidation and egress from the nucleus, the architecture of the nuclear lamina needs to be dissolved to allow the nucleocapsid to bud from the inner nuclear membrane into the perinuclear space (17). Indeed, infections by alpha- and betaherpesviruses induce changes in the nuclear lamina (44, 48, 51), whereas the status of the nuclear architecture during gammaherpesvirus infection remains unclear.
Interestingly, many of these nuclear changes induced by herpesvirus replication, such as cellular chromatin alternation and nuclear lamina disassembly, are quite similar to what happens during mitosis. Cellular Cdc2 (CDK1) is the mammalian cyclin-dependent kinase which controls multiple targets during mitosis. Cdc2 activation is mediated by a complex process which begins with an increase of cyclin B1 at S and G2 phases for Cdc2 nuclear targeting. The phosphorylation of Thr14 and Tyr15 on Cdc2 is then reversed by Cdc25C at the end of the G2 phase to turn on Cdc2 activity (33, 43, 46). Cdc2 kinase governs many cellular events at mitotic transition, including the breakdown of nuclear lamina, shutdown of transcription and DNA replication, and condensation of chromosomal DNA (14, 16, 19). Under normal cell cycle regulation, chromosome condensation ensures the appropriate segregation of genetic information during mitosis. Accumulating genetic and biochemical evidence suggests that condensin and DNA topoisomerase II (Topo II) play crucial roles in mitotic chromosome assembly and organization (9). Condensins are multisubunit protein complexes; they are composed of two structural maintenance of chromosome (SMC) subunits and three non-SMC regulatory subunits and play a central role in mitotic chromosome condensation and segregation (20). Condensins also participate in activating the DNA checkpoint, gene silencing, and the excision repair pathway (18). Human condensin is phosphorylated and activated by Cdc2 kinase in vitro and in vivo (27, 56). Topo II, which catalyzes a transient breakage and religation of double-stranded DNA, is the major constituent of the chromosome scaffold and essential for mitotic condensation (12, 58).
In HSV-1, UL13 kinase phosphorylates Us3 and subsequently regulates correct nuclear localization of UL34 and UL31, two HSV-1 gene products involved in nuclear egress (22, 28, 49). The possible contribution of EBV kinase to the virus replication-induced mitosis-like events is of special interest. EBV BGLF4 kinase is the only Ser/Thr kinase identified in EBV and is conserved in all herpesviruses. BGLF4 can phosphorylate viral DNA polymerase accessory factor EA-D (BMRF1), EBNA-2, EBNA-LP, and BZLF1 (3, 7, 23, 62). It colocalizes with the viral DNA replication compartment and is packaged into the mature virion as a tegument protein, suggesting that BGLF4 may be involved in virion maturation and packaging (3, 59). In addition, BGLF4 can phosphorylate the cellular translation elongation factor 1 delta and casein kinase IIβ at Cdc2 target sites (24). Thus, we wish to determine how BGLF4 affects the host cell during virus replication.
Previous studies of EBV reactivation mainly focused on the viral replication compartment and cellular environment, and little is known of how EBV reorganizes the nuclear architecture and chromatin territories. Here we demonstrate that cellular chromatin is condensed upon EBV replication and that EBV BGLF4 kinase alone can induce cellular DNA condensation through condensin phosphorylation and Topo II activation, coupled with multiple mitosis-like events which may facilitate EBV replication.
MATERIALS AND METHODS
Cell culture, virus induction, and transfection.
The HEK293T cell line was derived from human embryonic kidney cells with the expression of simian virus 40 T antigen. The HeLa cell line was derived from human cervical epithelial cells. EBV-negative cell line NPC-TW01 was established from a Taiwanese nasopharyngeal carcinoma (34), and the NA cell line was a recombinant Akata EBV-converted NPC-TW01 line (6). All cells were cultured in Dulbecco's modified Eagle's medium and supplemented with 8% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. Akata cells with or without EBV (55) were cultured in RPMI 1640 medium. For virus induction, NA cells were incubated with 40 ng 12-O-tetradecanoylphorbol-13-acetate-3 mM sodium butyrate (TPA/SB) or transfected with EBV Rta-expressing plasmid pRTS15 (50) using Lipofectamine (Invitrogen) in OptiMEM medium (GIBCO-BRL) according to the manufacturer's instructions. Akata cells (1 × 106 cells/ml) were induced with 0.5% (vol/vol) goat-anti human IgG antibody (Cappel).
pYPW17 and pYPW20 are plasmids expressing wild-type BGLF4 and K102I kinase mutant, respectively (59). GFP-BGLF4(pCPL4) was generated by cloning a BamHI-EBV BGLF4-EcoRI fragment from pYWP17 into the BglII-EcoRI cloning site of pEGFP-C1 (BD Biosciences). Flag-tagged HSV-1 UL13 (pTAG-UL13), human cytomegalovirus (HCMV) UL97(pTAG-UL97), and murine herpesvirus 68 (MHV68) ORF36 (pTAG-36) were generated independently by cloning BamHI-KpnI viral kinase gene fragments into pTAG-attR-C1 (Invitrogen) and were kindly provided by S. Hwang and R. Sun (University of California, Los Angeles).
Antibodies and indirect immunofluorescence.
Slide-cultured cells were transfected with plasmids expressing BGLF4, K102I, or vector pSG5. For BGLF4, lamin A/C, γ-tubulin, and actin fiber staining, the slides were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 24 h posttransfection at room temperature (RT) for 20 min, washed with PBS (145 mM NaCl, 1.56 mM Na2HPO4, 1 mM KH2PO4, pH 7.2), and permeabilized with 0.1% Triton X-100 at RT for 5 min. The slides were then incubated with anti-BGLF4 (2616 [59]), rabbit anti-BGLF4 serum, anti-lamin A/C (Santa Cruz), anti-γ-tubulin (Sigma), or anti-Flag (Sigma). For β- or F-actin staining, cells were fixed with methanol at −20°C for 30 min, washed with PBS, and incubated with anti-β-actin antibodies (Sigma) and rabbit anti-BGLF4 serum. All the primary antibodies were incubated with samples at 37°C for 1.5 h. After washes with PBS, slides were incubated with rhodamine-conjugated anti-mouse IgG (Cappel), fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibodies (Cappel), and/or FITC-phalloidin (10 μM; Sigma; filament actin binding toxin) at 37°C for 1 h. DNA was stained with Hoechst 33258 at RT for 30 s. The staining patterns were observed under fluorescence microscopy (Zeiss) or confocal microscopy (Leica). Approximately 150 to 300 cells were counted for each set of tests, and all the experiments were repeated independently at least twice. To detect the phosphorylation status of condensin, phosphospecific MPM-2 antibody (Upstate) was used in an immunoprecipitation-Western blot assay.
Coimmunoprecipitation and immunoblotting.
At 24 h posttransfection, HeLa cells (1 × 107) were harvested and disrupted in Nonidet P-40 (NP-40) lysis buffer (1% NP-40, 50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, and 1 mM Na3VO4) or CSK buffer (10 mM HEPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.25 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM NaF, 0.1 mM ATP) containing 0.3 M NaCl and 0.3% Triton X-100 (modified from reference 56). The cell lysates were separated by centrifugation for 10 min at 16,000 × g, and supernatant was recovered. Before immunoprecipitation, the lysate was incubated with 125 μl 20% protein A-Sepharose beads (Pharmacia) for 1 h at 4°C. To immunoprecipitate condensin complexes, lysates prepared in CSK buffer were incubated with human chromatin-associated protein E (hCAP-E) and G (hCAP-G) antibodies (1.5 μg each) at 4°C for 1 h. Protein A-Sepharose beads (125 μl at 20%) were added to pull down the immunocomplexes with rotation for 1 h at 4°C. The immunocomplexes were then washed extensively with CSK buffer, NP-40 lysis buffer, or cold PBS; disrupted in sodium dodecyl sulfate (SDS) sample buffer; and displayed on 8% SDS-polyacrylamide gels for immunoblotting detection.
For immunoblotting, proteins resolved by SDS-polyacrylamide gel electrophoresis (PAGE) were electrophoretically transferred onto Hybond-C Extra membranes (Amersham). The membranes were blocked, probed with different antibodies, and developed using an enhanced chemiluminescence kit (Amersham).
Chromosome spreads.
HeLa cells trypsinized at 24 h posttransfection or Akata cells induced with anti-IgG for 24 h were harvested and swollen in hypotonic solution (75 mM KCl, prewarmed to 37°C) for 20 min as previously described (5). Swollen cells were then washed in Carnoy's fixative (3:1 [vol/vol] methanol-acetic acid) overnight at 4°C. Fixed cells were dropped onto a coverslip covered with cold H2O or centrifuged onto slips at 2,000 rpm for 10 min in a cytocentrifuge (Cytospin 2; Thermo/Shandon). The chromosome spreads were fixed and processed for immunofluorescence.
DNA content analysis.
HeLa cells were transfected with pEGFP-C1 or pGFP-BGLF4, trypsinized at 24 h posttransfection, fixed with 1% paraformaldehyde on ice for 20 min and then with 75% ice-cold ethanol at −20°C overnight, washed with PBS, and resuspended in PBS containing 0.2% Triton X-100. RNase A was then added to a final concentration of 0.5%. Propidium iodide was then added to a 10-μg/ml concentration, and samples were incubated at RT for 30 min. DNA content was analyzed on a FACSCalibur cell sorter, and the data were processed with CellQuest software (BD Biosciences). The protein expression was confirmed using anti-green fluorescent protein (GFP) antibodies (BD Biosciences; JL-8).
Immune complex kinase assays.
HeLa cells were transfected with plasmid DNA and incubated with 5 μM roscovitine (Calbiochem) for 20 h. Cells were harvested and lysed in immunoprecipitation buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EGTA, 0.2 mM Na3VO4, 0.2 mM PMSF, 0.5% NP-40, and 50 mM NaF) containing a protease inhibitor cocktail (Roche). Immunoprecipitations were performed using anti-BGLF4 antibody; immunoprecipitates were collected by a brief centrifugation and washed extensively with immunoprecipitation buffer. Immunocomplexes were then washed once with kinase buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.5 mM dithiothreitol, 0.2 mM Na3VO4, and 0.1 mM NaF). For in vitro kinase assays, kinase buffer containing 25 μM ATP, 2.5 μCi [γ-32P]ATP, and 1 μg histone H1 (Calbiochem) was added to immunoprecipitates (10 μl) containing BGLF4 kinase with or without 5 μM roscovitine and incubated at 30°C for 30 min. After incubation, proteins resolved by SDS-PAGE were electrophoretically transferred onto Hybond-C Extra membranes (Amersham) and subjected to autoradiography.
Subcellular fractionation of CAPs.
Subcellular fractionation was basically performed as described previously (56). HeLa cells (5 × 105) were lysed in 200 μl CSK buffer containing 0.3% Triton X-100 at 4°C for 30 min. The soluble and insoluble fractions were separated by centrifugation for 10 min at 2,000 × g. The supernatant was recovered as the chromatin-unbound fraction (see Fig. 6A, Fr. 1). The insoluble fraction was suspended in 200 μl CSK buffer with 0.01 U/μl of DNase I, incubated at RT for 30 min to solubilize DNA, and clarified by centrifugation for 10 min at 17,500 × g. The supernatant was recovered as the DNase-extractable fraction (see Fig. 6A, Fr. 2). The pellet was resuspended in 200 μl CSK buffer with 2 M NaCl and 0.3% Triton X-100 and centrifuged for 10 min at 17,500 × g to remove DNA and DNA-associated proteins from the nucleus. The supernatant was saved as the high-salt-extractable fraction (see Fig. 6A, Fr. 3), and the pellets containing nuclear matrix were saved as the high-salt-resistant fraction (see Fig. 6A, Fr. 4). The DNase-extractable and high-salt-extractable fractions were considered chromatin-bound fractions.
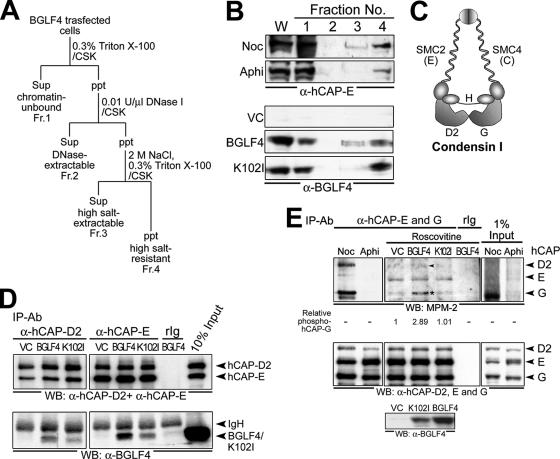
FIG. 6.
BGLF4 associates with cellular chromatin, interacts with condensin, and induces condensin phosphorylation. (A) Fractionation scheme of BGLF4-expressing cell lysate (see Materials and Methods for details). Sup, supernatant; ppt, precipitate; Fr., fraction. (B) Whole-cell extracts (W) and chromatin-unbound (Fr. 1), DNase-extractable (Fr. 2), high-salt-extractable (Fr. 3), and high-salt-resistant (Fr. 4) fractions prepared from vector (VC) or BGLF4- or K102I-expressing, nocodazole (Noc; 100 ng/ml)- or aphidicolin (Aphi; 1 μg/ml)-treated cells were subjected to immunoblot analysis with anti-hCAP-E (top) or anti-BGLF4 (bottom) antibody. (C) The composition of human condensin I complex includes core subunits SMC2 (hCAP-E) and SMC4 (hCAP-C) and regulatory subunits hCAP-D2, -H, and -G (adapted from the work of Hirano [20]). (D) Vector- or BGLF4 plasmid-transfected HeLa cell lysate was immunoprecipitated with anti-condensin subunit hCAP-D2 or -E antibody. The immunocomplexes were resolved by 8% SDS-PAGE and immunoblotted with antibodies to hCAP-D2 and hCAP-E (top) or BGLF4 (bottom). (E) BGLF4 induces condensin phosphorylation. Cell lysates harvested from 5-μM-roscovitine-treated HeLa cells expressing BGLF4 or vector only were immunoprecipitated by hCAP-E and -G antibodies. The phosphorylation status of condensin was detected by mitotic phosphoantibody MPM-2 (top), and the signal intensity of phosphorylated hCAP-G relative to that of vector control is indicated. Equal immunoprecipitation of condensin subunits was detected by anti-hCAP-D2, -E, and -G antibodies (middle). Expression levels of BGLF4 or K102I were detected with specific antibody (bottom). Noc and Aphi indicate nocodazole (100-ng/ml)- or aphidicolin (1-μg/ml)-treated cells, respectively. The arrowhead and the asterisk indicate phosphorylated condensin hCAP-D2 and -G subunits, respectively. IP, immunoprecipitation; Ab, antibody; WB, Western blot.
PFGE.
After transfection, HeLa cells were washed and resuspended in PBS at a concentration of 2 × 106 cells/ml. The cell suspension was mixed with an equal volume of 1.2% agarose and poured into sample holders. The agarose plugs (with 1 × 105 cells) were then transferred into 400 μl of sample lysis buffer (50 mM Tris-Cl, 10 mM EDTA, 0.5% SDS, and 10 mg/ml proteinase K in 50 mM CaCl2) and incubated at 50°C for 24 h. Electrophoresis was performed in 0.5× TBE buffer (45 mM Tris-HCl, pH 7.5, 45 mM boric acid, 1.25 mM EDTA) with a pulsed-field gel electrophoresis (PFGE) system (Bio-Rad) at 60 V, 14°C. The pulsed-field switch time was 15 s for 10 h, followed by a 60-s switch time for another 10 h. The DNA in the gel was then visualized by ethidium bromide staining.
DNA decatenation assay.
Topo II activity was assayed as previously described (52) with slight modification. HeLa cells (3 × 106) transfected with BGLF4, K102I, or vector were scraped from plates at 48 h posttransfection and swollen in cold TEMP buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 4 mM MgCl2, 0.5 mM PMSF) on ice for 10 min. The nuclear pellet was spun down at 1,500 × g for 10 min at 4°C, resuspended in 30 μl TMP buffer (TEMP buffer without MgCl2), mixed with 30 μl 1 M NaCl, vortexed, and left on ice for 60 min. After centrifugation at 15,000 × g for 15 min, the supernatant containing Topo II activity was aliquoted and the protein concentration was determined (modified from the protocol of the manufacturer [TopoGen, 2002]). Nuclear lysate (0.5 μg) was then incubated with 150 ng kinetoplast DNA (kDNA; TopoGen) in a final volume of 20 μl containing 10 mM Tris-HCl, pH 7.9, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 75 μg/ml bovine serum albumin, and 1 mM ATP at 30°C for 10, 30, or 60 min. Reaction mixtures were then quenched with 10 mM EDTA-0.1% SDS, and DNA products were resolved on 1% agarose-Tris-borate-EDTA gels and stained with ethidium bromide.
RESULTS
EBV reactivation induces cellular chromatin condensation and interchromosomal space enlargement.
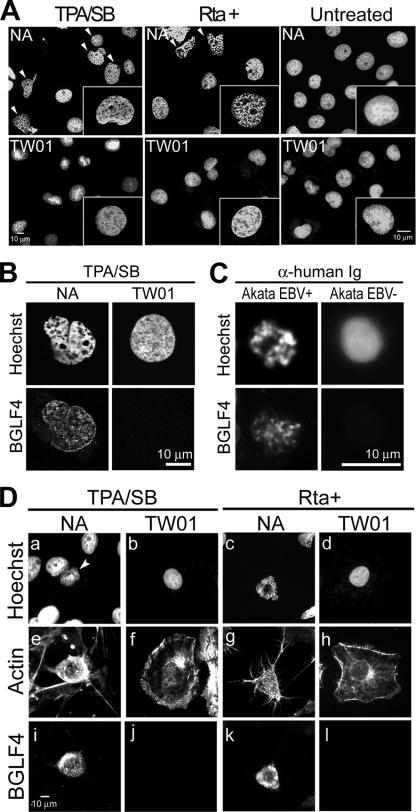
To observe possible changes in cellular chromatin architecture during EBV replication, we stained cellular chromatin with Hoechst 33258 in chemically reactivated EBV-positive NA cells and parental EBV-negative TW01 cells. Condensed chromatin with a partially irregular nuclear shape was consistently observed in >90% of NA cells but not in TW01 cells at 48 h postinduction (for each set of experiments, 300 cells were counted; Fig. 1A). To rule out possible effects of the chemical, we transiently expressed the EBV lytic transactivator Rta in NA and TW01 cells to induce virus replication. Dramatically condensed chromatin with enlarged interchromosomal space was observed in Rta-induced NA but not in TW01 cells or noninduced NA cells (Fig. 1A and 1B). Using BGLF4 expression as a marker for viral replication, similar condensed DNA also was observed in anti-IgG-induced Akata EBV-positive cells (95.2% of 150 BGLF4-positive cells) but not in EBV-negative Akata cells (4 to 8% in Akata EBV+ cells without anti-human Ig treatment or Akata EBV− cells displaying a mitotic DNA staining pattern). However, the cellular chromatin structure is more compact in Akata cells, probably due to the small nuclear size of lymphocytes. These observations suggest that EBV induces structural change of cellular chromatin during viral lytic replication.
FIG. 1.
EBV reactivation induces morphological changes in cellular chromatin. EBV-positive NA or EBV-negative TW01 cells were not treated, were treated with TPA/SB, or were transfected with a plasmid expressing Rta. At 24 h postinduction or -transfection, cells were fixed and cellular DNA was stained with Hoechst 33258. (A) Cellular chromatin morphology of cells replicating EBV (NA, TPA, and Rta+), TW01 cells, and untreated cells. Arrowheads indicate cells with an altered DNA staining pattern observed among NA but not among TW01 cells. For each set, 300 cells were counted, and the experiment was repeated independently more than three times. (B) Cellular chromatin morphology of NA cells replicating EBV. Chromatin architecture of NA or TW01 cells treated with TPA/SB was observed by confocal microscopy. Condensed chromosomes and enlarged interchromosomal space were observed in EBV-replicating NA but not TW01 cells. (C) Cellular chromatin morphology of EBV-replicating Akata cells. EBV-positive (EBV+) or -negative (EBV−) Akata cells were induced for lytic virus replication by 0.5% (vol/vol) anti-human IgG (α-human Ig) cross-linking. At 24 h post-Ig treatment, cells were fixed and chromosome spread assays were performed as described in Materials and Methods. DNA and BGLF4 were then detected by staining with Hoechst 33258 and BGLF4 antibody. For each set, 150 cells were counted and the experiment was repeated twice. (D) Cell morphology of chemically induced or Rta-transduced NA or TW01 cells was visualized by immunostaining of actin filaments (e to h). Nuclear DNA (a to d) and BGLF4 kinase (i to l) were detected by Hoechst 33258 staining and immunostaining to indicate cells replicating virus.
Although EBV-replicating cells show condensed cellular chromatin coupled with an irregular nuclear shape and enlarged interchromosomal space, no significant nuclear expansion was observed in these cells, in contrast to HSV-1 replication. Interestingly, when β-actin was stained to observe the morphology of induced cells, NA cells with condensed chromosomes showed a rounded-up morphology with contracted actin filament bundles, in contrast to the well-attached TW01 cells (Fig. 1D).
To search for a possible mechanism involved in these phenomena, we investigated the expression of several lytic viral genes and cellular proteins after viral replication (see Fig. S1 in the supplemental material). We found that the expression patterns of cellular Cdc2, cyclin B, and p21 were not significantly changed in TPA/SB-induced NA cells, compared with TW01 cells. Because BGLF4 potentially phosphorylated several proteins at the same target sites of cellular mitotic kinase Cdc2 (24), we suspected that BGLF4 kinase may be involved in inducing cellular chromatin condensation.
EBV BGLF4 kinase induces cellular chromosome condensation.
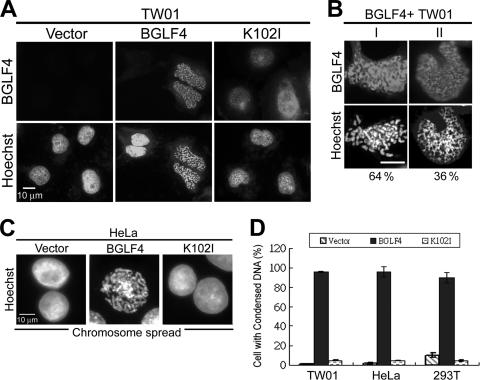
To determine whether BGLF4 can induce chromosome condensation, BGLF4 or K102I (kinase dead) was expressed in TW01 cells. The chromatin morphology and BGLF4 expression of transfected TW01 cells were examined by immunofluorescence assay. Surprisingly, the chromatin morphology of BGLF4-expressing cells showed a prophase-like individualized condensation pattern, whereas no morphological change in chromatin was observed in vector- or K102I-expressing cells (Fig. 2A). A similar individualized chromosome condensation also was observed in BGLF4- but not K102I-expressing HeLa and HEK293T cells (data not shown). Interestingly, the chromatin staining pattern in BGLF4-positive TW01 cells can be divided further into two subtypes, about 64% of cells with individualized condensed chromosomes and 36% with spongiform cellular chromatin (Fig. 2B; 300 cells/set were counted). The BGLF4-induced chromosome condensation in HeLa cells was further illustrated by chromosome spreading (Fig. 2C). The percentage of BGLF4-expressing cells with condensed chromosomes was then scored in three different cell lines (Fig. 2D). We found that almost every cell with BGLF4 showed condensed DNA staining (96% in TW01 and HeLa cells and 90% in HEK293T cells; 300 cells/set were counted). To avoid an artifact of overexpression, BGLF4 expression levels in anti-IgG-induced Akata cells, TPA/SB-induced NA cells, and transiently transfected cells were compared to confirm their equivalence (see Fig. S2 in the supplemental material). Taken together, we conclude that BGLF4 kinase induces cellular chromosome condensation in a kinase activity-dependent manner.
FIG. 2.
BGLF4 kinase induces cellular DNA condensation. (A) Slide-cultured TW01 cells were transfected with BGLF4- or K102I-expressing plasmids or pSG5. Cells were fixed at 24 h posttransfection and stained for BGLF4 and DNA as described in Materials and Methods. Condensed chromosomes were observed in cells expressing BGLF4 but not K102I. (B) Two types of cellular chromatin (I, with individualized condensed chromosomes; II, with sponge morphology chromatin) were observed in BGLF4-expressing TW01 cells. For each set, 300 cells were counted. (C) Chromosome spread assays of BGLF4-, K102I-, or vector-transfected HeLa cells were performed as described in Materials and Methods, and cells were stained with Hoechst 33258. (D) The percentage of 300 BGLF4-positive cells with condensed chromosomes was determined by fluorescence microscopy.
BGLF4 induces nuclear lamina disassembly and reorganization of the cytoskeleton but not centrosome polarization.
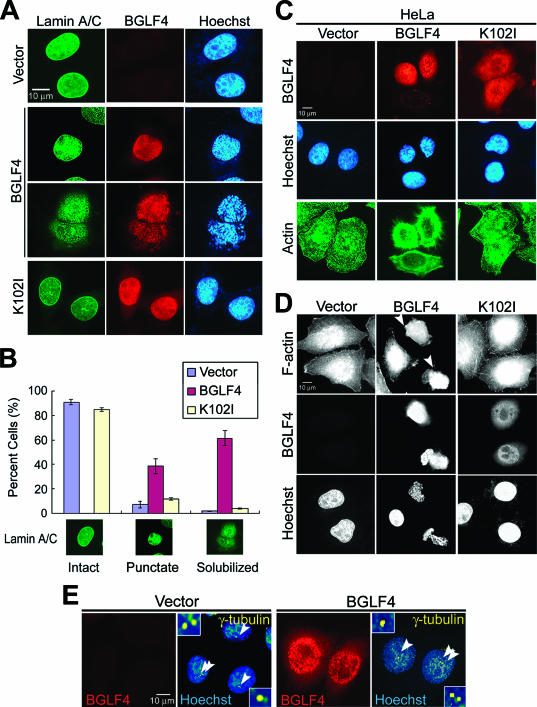
Chromosome condensation and rounded cell morphology usually occur at the mitotic phase, accompanied by disassembly of the nuclear lamina. We then explored whether BGLF4 induces other mitosis-like events. By staining of type A and C lamins, nuclear lamina disassembly was observed in 61.5% of BGLF4-expressing HeLa cells and another 38.5% of cells showed condensed punctate lamin staining, whereas nuclear lamina remained intact in 84.7% of cells expressing K102I (Fig. 3A). Cells were scored for intact, punctate, or solubilized nuclear lamin staining patterns (Fig. 3B), and almost every BGLF4-expressing cell (>99%) showed redistributed nuclear lamina. Similar to events in normal mitotic cells, the immunoblotting data indicated that lamin A/C underwent disassembly but not degradation (see Fig. S3B in the supplemental material). These results revealed that BGLF4 induces not only chromosome condensation but also redistribution of the nuclear lamina.
FIG. 3.
BGLF4 induces nuclear lamina disassembly and stress fiber reorganization accompanied by chromosome condensation. HeLa cells were transfected with BGLF4- or K102I-expressing plasmid or control vector. At 24 h posttransfection, cells were fixed with 4% paraformaldehyde and stained for BGLF4, lamin A/C, and DNA as described in Materials and Methods. (A) BGLF4-expressing cells showed diffused nuclear lamina coupling with condensed chromosomes. (B) The lamin A/C distribution pattern (∼300 cells/set) was scored as one of three types (intact, punctate, or solubilized patterns) as indicated. BGLF4-expressing cells show increased cell population with condensed punctate lamin staining (38.5% in BGLF4-, 6.9% in vector-, and 11.4% in K102I-expressing cells) or solubilized (61.5% in BGLF4-, 1.8% in vector-, and 3.9% in K102I-expressing cells) nuclear lamina. (C) HeLa cells were transfected with BGLF4- or K102I-expressing or control vector; fixed with methanol; and stained for BGLF4, actin, and DNA. Rounded cell morphology with rearranged actin filament was observed in BGLF4-expressing cells. (D) F-actin and DNA staining patterns of HeLa cells transfected with the indicated plasmids. F-actin was detected by FITC-conjugated phalloidin. Arrowheads indicate BGLF4-expressing cells with rearranged actin filaments. (E) Staining patterns of γ-tubulin, BGLF4, and DNA in transfected HeLa cells. Arrowheads indicate the γ-tubulin representing centrosome localization. BGLF4-expressing cells (∼95% of 150 cells) show nonseparated centrosomes similar to those in vector-transfected cells. However, a slight change in the morphology of the centrosome was observed in cells expressing BGLF4 (∼22% of 150 cells).
To explore the role of BGLF4 in the regulation of cytoskeleton rearrangement which may lead to a subsequent change of cell morphology, we stained β-actin to indicate the cytoskeleton and found that cells expressing BGLF4 but not K102I showed contracted actin-filament bundles (Fig. 3C). Simultaneously, FITC-phalloidin, an actin-filament binding toxin, was used to detect F-actin stress fibers in BGLF4-positive cells (Fig. 3D). Heavily condensed and rearranged stress fibers were observed in cells expressing BGLF4 but not K102I, indicating that expression of BGLF4 can induce rearrangement of actin filaments directly or indirectly. Furthermore, redistribution of α-tubulin, a major component of microtubules, also was observed in BGLF4-expressing cells (data not shown).
In normal cell cycle progression, the separation (polarization) of duplicated centrosomes should couple with chromosome condensation, nuclear lamina disassembly, and cell rounding during mitosis. We detected centrosomes by immunostaining of γ-tubulin, which is the major component of microtubule-organizing centers and can indicate the position of centrosomes. Even though some cells (∼22%) showed slightly irregular centrosome morphology, nonpolarized centrosomes (number 1 or 2, ∼95% in 150 cells) were observed in BGLF4-expressing cells (Fig. 3E). This indicates that BGLF4 has no effect on centrosome replication and separation. Taken together, data here suggest that BGLF4 can induce several coupled mitosis-like events, including chromosome condensation, nuclear lamina disassembly, and cell rounding, but these are not coordinated with centrosome separation.
BGLF4 induces premature chromosome condensation independently of G2/M accumulation and Cdc2 signaling.
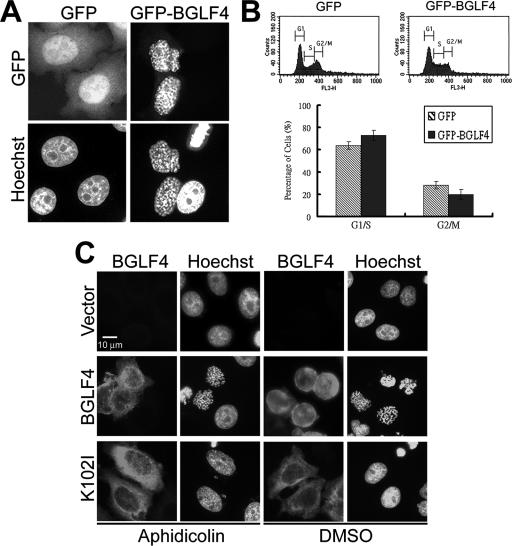
In normal cell cycle progression, chromosome condensation is triggered by Cdc2 kinase at entry to mitosis (20). We wondered whether BGLF4 induces chromosome condensation by increasing mitotic cells or through the Cdc2-mediated signaling pathway. To monitor DNA content, it was first confirmed that GFP-BGLF4 but not GFP vector was able to induce chromosome condensation in HeLa cells (Fig. 4A). After cells with green fluorescence were gated, the DNA content of transfected cells was analyzed by flow cytometry. In asynchronized HeLa cells expressing GFP-BGLF4, the 2N cell population (G1/S border) increased approximately 10% whereas the cell population with 4N DNA (G2/M border) was slightly reduced (Fig. 4B). To test whether BGLF4 induces premature chromosome condensation independently of DNA replication, HeLa cells transiently expressing BGLF4 were synchronized at S phase using the DNA polymerase inhibitor aphidicolin. The efficacy of aphidicolin treatment was confirmed by the accumulation of phosphorylated Cdc2 (Fig. 5A). Chromosome condensation was observed in cells expressing BGLF4 but not K102I (Fig. 4C). These results indicate that BGLF4 induces premature chromosome condensation independently of accumulation of mitotic (4N) cells.
FIG. 4.
BGLF4 induces chromosome condensation independently of G2/M accumulation and DNA replication. HeLa cells were transfected with GFP-BGLF4 (pCPL4) or pEGFP-C1 and incubated for 24 h. (A) Transfected cells were stained for GFP and cellular DNA. GFP-BGLF4 but not GFP vector induces condensed chromosomes. (B) The DNA content and cell cycle distribution of gated GFP- or GFP-BGLF4-expressing cells were detected on a FACSCalibur cell sorter and analyzed using CellQuest software as described in Materials and Methods. No significant accumulation in G2/M (4N) cell population was observed in GFP-BGLF4-expressing cells. (C) BGLF4 induces chromosome condensation independently of DNA replication. Slide-cultured HeLa cells were transfected with BGLF4, K102I, or control vector and synchronized at G1/S phase by 1 μM aphidicolin treatment for 20 h. Cells were then fixed and stained for BGLF4 and DNA at 24 h posttransfection. Dimethyl sulfoxide (DMSO) served as a solvent control. Nocodazole-treated cells served as an MPM-2 detection control.
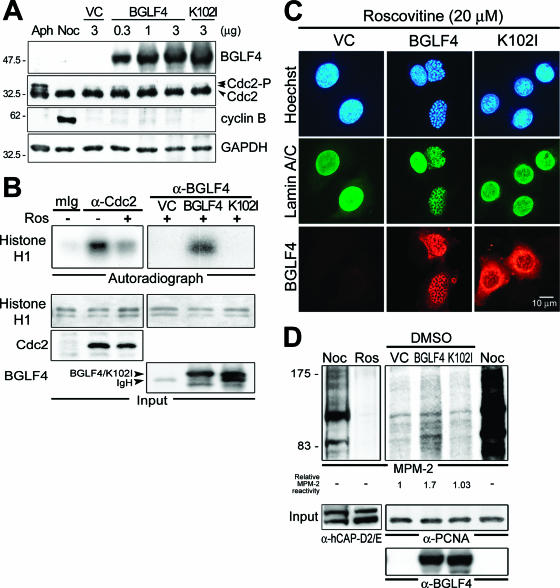
FIG. 5.
BGLF4 induces chromosome condensation independently of Cdc2 signaling. (A) Cdc2 and cyclin B protein expression in BGLF4-expressing cells. HeLa cells were transfected with BGLF4, K102I, or control vector (VC). At 24 h posttransfection, Cdc2 and cyclin B expression was detected using anti-Cdc2 and anti-cyclin B1 antibodies, respectively. No cyclin B accumulation or change in phosphorylation pattern of Cdc2 was observed, suggesting that Cdc2 activity may not be activated in cells expressing BGLF4. HeLa cells treated with 1 μM aphidicolin (Aph) or 50 ng/ml nocodazole (Noc) served as S- and M-phase controls, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Numbers at left are molecular masses in kilodaltons. (B) Immune complex kinase assays of Cdc2 and BGLF4. The kinase activity of immunoprecipitated Cdc2, BGLF4, or K102I under roscovitine (Ros; 5 μM) treatment was assayed as described in Materials and Methods using histone H1 (1 μg) as kinase substrate. Input protein was detected by Cdc2, BGLF4 antibody, or silver staining (histone H1), respectively. (C) Slide-cultured HeLa cells were transfected with BGLF4, K102I, or control vector (VC) and incubated in the presence of the Cdc2 inhibitor roscovitine (20 μM) for 20 h. Cells were then fixed and stained for BGLF4, lamin A/C, and DNA as described in Materials and Methods. (D) BGLF4 induces multiple protein phosphorylation on MPM-2 epitope. HeLa cells were treated with nocodazole (Noc) or roscovitine (Ros; 5 μM) for 20 h. The protein phosphorylation pattern was detected by MPM-2 antibody and then quantified by ImageQuant software. Significantly reduced MPM-2 phosphorylation was detected in roscovitine-treated cells, and increased MPM-2 signal (1.7-fold over that of vector control) was detected in BGLF4-expressing cells. Condensin subunit hCAP-D2 and -E or PCNA served as a loading control. DMSO, dimethyl sulfoxide.
Because several coupled cellular events are regulated by Cdc2 at entry to mitosis (11, 19, 60), we determined whether BGLF4 induces chromosome condensation through activation of Cdc2. Expression patterns of Cdc2 and cyclin B were detected in cells expressing BGLF4 and K102I, and no significant difference was observed (Fig. 5A). Furthermore, a Cdc2-specific inhibitor, roscovitine, was used to repress endogenous Cdc2 activity in BGLF4-expressing cells. We found that roscovitine can significantly repress the activity of immunoprecipitated Cdc2 but not the kinase activity of BGLF4 on histone H1 (Fig. 5B). Even with the treatment of a high concentration (20 μM) of roscovitine, chromosome condensation and nuclear lamin disassembly were induced by BGLF4, suggesting that BGLF4 induces chromosome condensation independently of Cdc2-mediated signaling (Fig. 5C). Interestingly, when we detected overall Ser/Thr phosphorylation patterns by using the phosphospecific antibody MPM-2, which recognizes a class of mitosis-specific phosphoproteins (10), relatively stronger signals were observed in BGLF4-expressing cells (Fig. 5D, 1.7-fold over that of vector-transfected cells), suggesting that BGLF4 may induce phosphorylation on multiple MPM-2 epitopes.
BGLF4 associates with cellular chromatin, interacts with condensin, and induces condensin phosphorylation.
Chromosome condensation is a multiple-step process mediated by the coordination of cellular chromatin-associated factors, condensin complexes, and Topo IIα (reviewed in references 15 and 20). We then examined whether BGLF4 associates with chromatin and interacts with condensin or Topo IIα. BGLF4-expressing cells were treated with CSK buffer containing Triton X-100, DNase I, or 2 M NaCl (Fig. 6A). Proteins not bound to chromatin were eluted by CSK buffer containing Triton X-100 (Fr. 1), and proteins bound to chromatin were recovered from DNase I-(Fr. 2) or high-salt-treated (Fr. 3) fractions. As described previously, condensin subunit hCAP-E can be found in the DNase I- and high-salt-treated fractions of nocodazole-treated cells (56). BGLF4 and K102I were detected in CSK buffer containing Triton X-100 and high-salt-extractable fractions, indicating that BGLF4 is a component of the nuclear matrix and can associate with chromatin structure (fractions 1 and 3 in Fig. 6B).
Human condensin I is a five-subunit protein complex, including core subunits SMC2 (hCAP-E) and SMC4 (hCAP-C) and regulatory subunits hCAP-D2, -H, and -G (Fig. 6C). Because Cdc2-phosphorylated condensin complex shows enhanced supercoiling activity and knotting activity in vitro (26, 56), we investigated the possible interaction between BGLF4 and condensin complexes by using hCAP-D2 or -E antibody. As expected, a small portion of BGLF4 and K102I could be coimmunoprecipitated with hCAP-D2 regulatory subunit and hCAP-E core subunit (Fig. 6D). Therefore, we tested the ability of BGLF4 to induce condensin phosphorylation by using phosphospecific antibody MPM-2 (10). In the presence of roscovitine, phosphorylation of hCAP-G was enhanced 2.89-fold by BGLF4 compared to that of vector control (asterisk in Fig. 6E) and slightly enhanced phosphorylation of hCAP-D2 (arrowhead in Fig. 6E) was observed in cells expressing BGLF4 but not K102I or vector. Because phosphorylation of condensin on MPM-2 epitopes increases its DNA-supercoiling activity (56), results here suggest that BGLF4 associates with cellular chromatin and induces chromosome condensation through condensin phosphorylation independently of Cdc2.
BGLF4 stimulates Topo II activity.
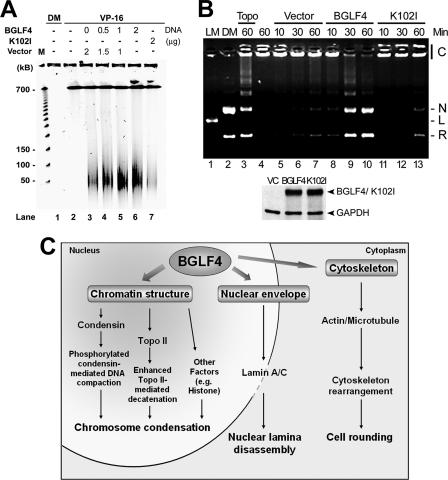
During mitosis, Cdc2 can associate physically with Topo IIα and enhance its decatenation activity (14), which is important for chromosome condensation and segregation (1, 53). Covalent binding between Topo II and DNA can be detected by the addition of a Topo II inhibitor, VP-16, followed by treatment with proteinase K to reveal the Topo II-mediated double-strand breaks that usually generate approximately 50- to 150-kb DNA fragments (14). The effect of BGLF4 on Topo II-DNA interaction in the presence of the Topo II inhibitor VP-16 was revealed by PFGE. BGLF4 but not K102I stimulated the formation of high-molecular-weight DNA fragments in a dose-dependent manner (Fig. 7A). To determine whether Topo II activity could be enhanced by BGLF4 kinase, nuclear extracts were prepared from BGLF4-expressing cells and incubated with the Topo II-specific substrate kDNA. Within 30 min, kDNA was effectively decatenated by a nuclear extract from cells transfected with BGLF4 but not from vector- or K102I-transfected cells (Fig. 7B). Taken together, BGLF4 can stimulate Topo II decatenation activity and Topo II-DNA interaction. Although we do not exactly know how BLGF4 stimulates Topo II, the elevated Topo II activity may coordinate with activated condensin complexes to induce chromosome condensation (Fig. 7C).
FIG. 7.
BGLF4 stimulates Topo IIα activity. (A) BGLF4 stimulates Topo II-DNA interaction. Transfected HeLa cells were incubated with 25 μM VP-16 or dimethyl sulfoxide (DM) control. At 24 h posttransfection, cells were subjected to PFGE to resolve DNA between 50 and 150 kb as described in Materials and Methods. The DNA fragment was stained with ethidium bromide. Lane 1, dimethyl sulfoxide treatment. Lane 2, mock transfection control. (B) Nuclear extracts from HeLa cells transfected with BGLF4, K102I plasmid, or vector were incubated with kDNA at 30°C for the time indicated to detect the decatenation activity. The DNA product was displayed on an agarose gel containing ethidium bromide. kDNA incubated with purified Topo II (1 U) served as a positive control (lane 3). Similar DNA patterns were reproducibly observed in two separate experiments. Lane 1, LM, XhoI-cut linear DNA marker; lane 2, DM, Topo II-decatenated DNA marker; lane 4, reaction buffer with kDNA only. C, catenated kDNA; N, nicked DNA; L, linear DNA; R, relaxed DNA. (Bottom) Similar protein expression of BGLF4 and K102I was confirmed by immunoblotting. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Summary of BGLF4-induced multiple premature mitotic events observed in this study. BGLF4 induces chromosome condensation by inducing condensin (hCAP-G) phosphorylation and enhancing Topo II-mediated DNA decatenation activity. Simultaneously, nuclear lamina disassembly and cytoskeleton rearrangement, which may subsequently induce rounded cell morphology, occur with condensed chromosomes.
Gammaherpesvirus kinases can induce chromosome condensation.
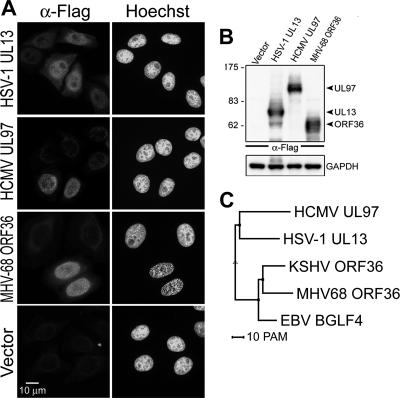
Morphological changes of the nucleus and cellular chromatin compression were reported in HSV-1-replicating cells (42), but the ability of HSV-1 UL13 kinase to induce these phenomena was never investigated. We were curious to know whether all UL13 homologues share the same ability with EBV BGLF4 to induce chromosome condensation. To this end, Flag-tagged HSV-1 UL13, HCMV UL97, and MHV68 ORF36 were expressed in HeLa cells for detecting the cellular chromatin staining pattern (Fig. 8A and B). We found that MHV68 ORF36 can dramatically induce chromosome condensation (93.2%) similar to that observed in BGLF4-expressing cells. Some HSV-1 UL13-expressing cells show a partial, uneven nuclear staining pattern, and no nuclear morphological change was observed in cells expressing HCMV UL97. Interestingly, phylogenic analysis and protein sequence alignment also indicate that gammaherpesvirus kinases EBV BGLF4, Kaposi's sarcoma-associated herpesvirus ORF36, and MHV68 ORF36 share high sequence similarity, whereas UL13 and UL97 contain extended amino-terminal ends (Fig. 8C; see also Fig. S4 in the supplemental material). Taken together, these observations indicate that gammaherpesvirus kinases EBV BGLF4 and MHV68 ORF36 have an ability to regulate the nuclear environment through inducing host chromatin condensation.
FIG. 8.
MHV68 ORF36 kinase induces chromosome condensation in transiently transfected HeLa cells. (A) Slide-cultured HeLa cells were transfected with Flag-tagged HSV-1 UL13-, HCMV UL97-, or MHV68 ORF36-expressing plasmids or control vector. At 24 h posttransfection, cells were fixed and stained for Flag-tagged protein as described in Materials and Methods. Cellular DNA was stained with Hoechst 33258. Flag-tagged protein-positive cells (∼200 cells/set) with condensed chromosomes were counted. Chromosome condensation was observed in 93.2% of cells expressing Flag-tagged MHV68 ORF36. (B) Flag-tagged protein expression was confirmed by immunoblotting using anti-Flag antibodies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. Numbers at left are molecular masses in kilodaltons. (C) Phylogenetic analysis of HSV-1 UL13, HCMV UL97, EBV BGLF4, Kaposi's sarcoma-associated herpesvirus (KSHV) ORF36, and MHV68 ORF36. PAM (point-accepted mutations), 1 unit of evolution as the amount of evolution that will change 1 in 100 amino acids on average.
DISCUSSION
Replication of DNA viruses must overcome several cellular restrictions, such as competing with host DNA for replication resources and nuclear space, and the resultant nucleocapsids need to exit from the nuclear envelope. Upon the induction of lytic replication, EBV creates an environment with high levels of S-phase cyclin-dependent kinase activities whereby cellular DNA replication is inhibited (30, 31), being partly regulated by phosphorylation of cellular MCM4 complexes by BGLF4 kinase (29). Here, we reveal a novel mechanism by which BGLF4 kinase remodels the cellular chromatin architecture and induces several mitosis-like events that may facilitate virus replication.
EBV replication induces changes in cellular chromatin architecture.
We observed in EBV-replicating NPC cells that condensation of the cellular chromatin was coupled with a rounded morphology (Fig. 1). A similar phenomenon also was observed in anti-IgG-reactivated EBV-positive Akata cells, suggesting that chromatin alteration during viral replication is similar to that observed in HSV-1-replicating cells but with some differences. Cells replicating HSV-1 usually show a progressive expansion of the nucleus and viral replication compartment accompanied by “compression and marginalization of host chromatin” (42). Here, no significant nuclear expansion was observed in cells replicating EBV. It seems that these two viruses employ different mechanisms to overcome the nuclear space limitation for viral DNA replication. In HSV replication, nuclear expansion can provide additional space for viral replication compartments; thus, the cellular chromatin does not need to be heavily condensed. In contrast, the EBV replication compartment seems to be more dispersed and distributed in the interchromosomal space (Fig. 1B); thus, chromosome compaction may provide enough space for the correct assembly of the EBV replication compartment.
BGLF4 induces cellular chromatin architecture change.
In searching for EBV gene products which might be responsible for structural changes in chromatin, we demonstrated that BGLF4 regulates chromosome condensation and other mitosis-like events, such as lamin disassembly and rearrangement of the cytoskeleton, in a manner dependent on kinase activity (Fig. 2 and 3). Interestingly, chromatin staining patterns induced by BGLF4 were slightly different from those of EBV-replicating cells. BGLF4 alone induced a prophase-like chromosome condensation; whereas a spongy DNA staining pattern without heavy compaction was observed in most BGLF4-positive cells replicating EBV (Fig. 1). It is possible that the BGLF4 protein expression level in a single cell is higher in transient transfection than in reactivated EBV-positive cells, even though we showed by Western blotting that the amounts of plasmid used in transfection were relevant to EBV-replicating cells (see Fig. S2 in the supplemental material). Alternatively, other viral products or replication compartments may coordinate with BGLF4 to induce chromatin structural changes in the complete virus system. Our finding here suggests that BGLF4 serves as the key player that induces chromosome condensation during EBV replication.
BGLF4 induces chromosome condensation through Cdc2 mimicry.
Chromosome condensation is a complicated process controlled predominantly by condensin complexes and Topo II. Here, we demonstrate that BGLF4 associates with cellular chromatin, interacts with the chromosome-remodeling factors condensin complexes, and induces condensin phosphorylation in the presence of the Cdc2 inhibitor roscovitine (Fig. 6). Using the mitosis-specific MPM-2 antibody, we show that BGLF4 induces condensin phosphorylation at the Cdc2 target motifs (56), suggesting that BGLF4 is a potential novel viral kinase that phosphorylates condensin and also might induce chromosome condensation by mimicking Cdc2. Searching potential phosphorylation sites, we found that the condensin subunit hCAP-G contains three major phosphorylation clusters in its carboxyl terminus (amino acids 660 to 765, 777 to 861, and 915 to 1015). Because the precise sites of Cdc2-induced phosphorylation on hCAP-G have not been mapped, we are interested in identifying BGLF4 target sites. However, no phosphorylated signal was detected using immunoprecipitated BGLF4 and glutathione S-transferase-hCAP-G (amino acids 915 to 1015) in our preliminary in vitro kinase assay, suggesting that BGLF4 and/or Cdc2 may phosphorylate other sites to activate condensin complexes. Currently we are investigating Thr322, Ser841, and Ser844 on hCAP-G for possible BGLF4-directed phosphorylation. On the other hand, it is known that EBV lytic replication elicits DNA checkpoint signal transduction (32), and condensin also participates in the DNA repair pathway (18). Further study will be needed to dissect the involvement of condensin in EBV-induced DNA checkpoint activation.
Topoisomerases are highly specialized enzymes involved in DNA metabolism, including replication, transcription, recombination, condensation, and segregation (8). Specifically, Topo IIα has been implicated as a critical enzyme involved in chromosome condensation and segregation (1, 53). As demonstrated by inhibition experiments, Topo IIα is required for HSV-1 and EBV replication (2, 25), possibly in untangling concatemeric viral DNA. Here, we demonstrate that BGLF4 not only enhances Topo II-DNA interaction but also stimulates Topo II activity in a kinase activity-dependent manner (Fig. 7A and B). Because BGLF4 is a protein kinase, it is possible that it may regulate Topo II activity through phosphorylation. However, the effect of phosphorylation on Topo II activity remains controversial. Topo II has been shown to be phosphorylated by Cdc2, protein kinase C (PKC), and mitogen-activated protein kinase in a cell cycle-dependent manner and by CKII in a cell cycle-independent manner (reviewed in reference 4). Even though it was shown that cellular kinases such as extracellular signal-regulated kinase 2 (ERK2) or Cdc2 activate Topo II independently of phosphorylation (14, 52), a recent study clearly indicated that the phosphorylation status of Topo II may affect its interaction with other proteins, such as BRCA1 (35). Therefore, further study will be required to reveal whether BGLF4 stimulates Topo II activity through direct phosphorylation or by regulating the interaction between Topo II and other factors.
In addition to condensin and Topo II, some studies suggested that the phosphorylation of histone H1 and/or H3 may be important for chromosome condensation (39, 45). Here, no dramatic histone H3 Ser10 phosphorylation, similar to that in mitotic cells, was detected in BGLF4-expressing NA cells, although partial BGLF4 and phosphorylated histone H3 colocalization was detected (data not shown). This suggests that BGLF4 may not contribute to histone H3-associated chromatin compaction. Conversely, histone H1 was phosphorylated by BGLF4 in vitro (Fig. 5B), suggesting that it may be a BGLF4 substrate which contributes to premature chromosome condensation. Taken together, these results suggest that condensin activation, Topo II-DNA interaction, and histone H1 phosphorylation may participate coordinately in BGLF4-induced chromosome condensation (Fig. 7C).
BGLF4-induced premature mitotic events may facilitate virus replication and maturation.
The cellular replication origin binding MCM4-MCM6-MCM7 complex was shown recently to be phosphorylated by BGLF4 during lytic replication of EBV (29). That phosphorylation at MCM4 inactivates its helicase activity for the initiation of cellular DNA replication illustrates how a virus can block host DNA replication in an S-like cellular environment. In our studies using bromodeoxyuridine or thymidine incorporation assays, similar repression of cellular DNA replication was observed in BGLF4-expressing cells (data not shown). It is known that condensed chromosomes show reduced efficiency for transcription and DNA replication during mitosis (16). We suggest that BGLF4-induced chromosome condensation also may contribute to the suppression of cellular DNA replication and transcription in cells replicating EBV. In addition, the unusual cellular chromatin compression can provide more interchromosomal space for virus replication. Thus, BGLF4 seems to be a major player in ensuring that the virus can take over the cellular DNA replication machinery.
Furthermore, we consider that the BGLF4-induced premature mitosis-like events, including redistribution of the nuclear lamina and actin fiber rearrangement, may facilitate virus maturation during the late phase of virus replication (Fig. 7C). When a DNA virus genome is packaged and egresses from nucleus to cytoplasm, it faces several barriers similar to the cellular genome in the partition of genetic materials at mitosis. From our earlier EBV microarray analysis, we found that BGLF4 is expressed during EBV reactivation in the second cluster of early genes, after the first cluster of genes such as those for DNA polymerase and processivity factor (37). In terms of cell cycle control, BGLF4 expression is very similar to the activation of Cdc2 after cellular DNA replication in late S phase. Although it is not sufficient to know whether BGLF4 also contributes to the compaction of the viral genome, accumulating evidence implies possible roles for herpesviral kinases in inducing some phenomenon resembling premature mitotic events, such as nuclear lamina redistribution and cytoskeleton rearrangement. HSV-1 induces reorganization of the nuclear lamina and promotes late virus maturation through viral UL13 or recruited cellular PKC (22, 47). Similarly, murine CMV and HCMV replication induces reorganization of the nuclear lamina by recruitment of cellular PKC or viral UL97 kinase during viral replication (40, 44). Hence, we suggest that BGLF4 may function in a Cdc2-mimicking mechanism to help virus encapsidation and egress.
Possible effects of BGLF4 on EBV pathogenesis.
Here, we found that BGLF4 induces “premature chromosome condensation” independently of Cdc2 signaling and cell cycle regulation (Fig. 4 and 5). Disassembly of nuclear lamina and rearrangement of the cytoskeleton accompanied by premature chromosome condensation also were observed in cells expressing constitutive Cdc2 kinase (21). Our results suggest that BGLF4 is a potential novel kinase which can trigger mitosis-like events similarly to constitutively active Cdc2. We found that most cells with BGLF4 expression and condensed chromosomes do not progress to division. This is reasonable because cell division would be detrimental to the virus while viral genomes are being replicated and encapsidated. Interestingly, although BGLF4-expressing cells showed a lower colony-forming ability than control cells, there are still some colonies formed by cells expressing BGLF4 after selection for 3 months (data not shown). This suggests that some cells may escape the consequences induced by BGLF4 and grow continuously. However, because premature condensation is correlated with induction of DNA breaks at replicating chromosome-bearing common fragile sites (13), it will be interesting to evaluate whether constitutive expression of a low level of BGLF4 may affect host genome stability.
Kinase-induced cellular chromatin architecture change is conserved in gammaherpesviruses.
Presumably all herpesviruses need to overcome similar barriers during their replication. In this study, we found that EBV and MHV68 kinases induce dramatic chromatin architecture change and cells expressing HSV-1 UL13 show spongy DNA staining morphology, whereas nuclear DNA remains homogenous in cells expressing HCMV UL97 (Fig. 8A). Because nuclear expansion was observed in cells replicating HSV-1 along with assembly of the viral replication compartment and compression and marginalization of host chromatin (42), it is possible that a more compact structural change is required in EBV- and MHV68-replicating cells to provide enough nuclear space for the viral compartments.
Multiple sequence alignment and phylogenic analysis indicate that HSV-1 UL13 and HCMV UL97 contain extended amino-terminal sequences (see Fig. S4 in the supplemental material; Fig. 8C). Overall, UL13 homologues of alpha-, beta-, and gammaherpesviruses appear to govern important nuclear functions during virus replication, with different pathways involved. Here we demonstrate that EBV BGLF4, but not UL13 or UL97, is able to dissolve and internalize nuclear lamina. It is possible that HSV UL13 and HCMV UL97 may cooperate with other viral or modified cellular factors to induce structural changes in chromatin that benefit virus replication. Alternatively, we could not exclude a minor possibility that Flag tagging may destroy any activity of UL13 or UL97 but not gammaherpesvirus kinases.
Taken together, we demonstrate that BGLF4 kinase is the key player that induces premature chromosome condensation and mitosis-like events in lytic EBV-replicating cells. BGLF4 induces these premature mitosis-like events independently of Cdc2-mediated signaling or cell cycle progression. From our investigation of the mechanisms involved in BGLF4-induced chromosome condensation, we suggest that phosphorylation of condensin and Topo II activity regulated by the viral kinase are very likely to contribute to changes in cellular DNA architecture. The ability to induce chromosome condensation is conserved in gammaherpesvirus kinases, whereas the kinases of other herpesviruses seem to utilize other strategies to ensure nuclear space for successful viral replication.
Supplementary Material
Acknowledgments
We thank Ren Sun and Seungmin Hwang at the University of California, Los Angeles, for Flag-tagged herpesvirus kinase-expressing vectors. We also thank Tsai-Kun Li and Ching-Hwa Tsai in the Department of Microbiology and Zee-Fen Chang and Jin-Yuh Shew in the Department of Biochemistry, NTUCM, for PFGE techniques and helpful discussion. We also thank Tim J. Harrison of University College London for critical reading and modification of the manuscript.
This study was supported by grants NHRI-EX94-9313BI and NHRI-EX95-9313BI from the National Health Research Institutes, Taiwan.
Footnotes
Published ahead of print on 14 March 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, Y., M. Luke, and U. K. Laemmli. 1991. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell 64:137-148. [DOI] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2003. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase II alpha for post-DNA synthesis expression of late genes. Proc. Natl. Acad. Sci. USA 100:4825-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 80:5125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, C. A., and K. L. Marsh. 1998. Eukaryotic DNA topoisomerase II beta. Bioessays 20:215-226. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, A. J., and A. C. Porter. 2004. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIα mutant human cell line. Mol. Biol. Cell 15:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y., C. H. Tung, Y. T. Huang, J. Lu, J. Y. Chen, and C. H. Tsai. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J. Virol. 73:8857-8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 74:3093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortes, F., N. Pastor, S. Mateos, and I. Dominguez. 2003. Roles of DNA topoisomerases in chromosome segregation and mitosis. Mutat. Res. 543:59-66. [DOI] [PubMed] [Google Scholar]
- 9.Cuvier, O., and T. Hirano. 2003. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 160:645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, F. M., T. Y. Tsao, S. K. Fowler, and P. N. Rao. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA 80:2926-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessev, G., C. Iovcheva-Dessev, J. R. Bischoff, D. Beach, and R. Goldman. 1991. A complex containing p34cdc2 and cyclin B phosphorylates the nuclear lamin and disassembles nuclei of clam oocytes in vitro. J. Cell Biol. 112:523-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earnshaw, W. C., B. Halligan, C. A. Cooke, M. M. Heck, and L. F. Liu. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Achkar, E., M. Gerbault-Seureau, M. Muleris, B. Dutrillaux, and M. Debatisse. 2005. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc. Natl. Acad. Sci. USA 102:18069-18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escargueil, A. E., S. Y. Plisov, A. Skladanowski, A. Borgne, L. Meijer, G. J. Gorbsky, and A. K. Larsen. 2001. Recruitment of cdc2 kinase by DNA topoisomerase II is coupled to chromatin remodeling. FASEB J. 15:2288-2290. [DOI] [PubMed] [Google Scholar]
- 15.Gassmann, R., P. Vagnarelli, D. Hudson, and W. C. Earnshaw. 2004. Mitotic chromosome formation and the condensin paradox. Exp. Cell Res. 296:35-42. [DOI] [PubMed] [Google Scholar]
- 16.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 17.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagstrom, K. A., and B. J. Meyer. 2003. Condensin and cohesin: more than chromosome compactor and glue. Nat. Rev. Genet. 4:520-534. [DOI] [PubMed] [Google Scholar]
- 19.Heald, R., and F. McKeon. 1990. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61:579-589. [DOI] [PubMed] [Google Scholar]
- 20.Hirano, T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265-R275. [DOI] [PubMed] [Google Scholar]
- 21.Jin, P., S. Hardy, and D. O. Morgan. 1998. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 141:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 84:3381-3392. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawanishi, M. 1993. Topoisomerase I and II activities are required for Epstein-Barr virus replication. J. Gen. Virol. 74:2263-2268. [DOI] [PubMed] [Google Scholar]
- 26.Kimura, K., O. Cuvier, and T. Hirano. 2001. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 276:5417-5420. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, K., M. Hirano, R. Kobayashi, and T. Hirano. 1998. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282:487-490. [DOI] [PubMed] [Google Scholar]
- 28.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudoh, A., T. Daikoku, Y. Ishimi, Y. Kawaguchi, N. Shirata, S. Iwahori, H. Isomura, and T. Tsurumi. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 80:10064-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudoh, A., T. Daikoku, Y. Sugaya, H. Isomura, M. Fujita, T. Kiyono, Y. Nishiyama, and T. Tsurumi. 2004. Inhibition of S-phase cyclin-dependent kinase activity blocks expression of Epstein-Barr virus immediate-early and early genes, preventing viral lytic replication. J. Virol. 78:104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudoh, A., M. Fujita, T. Kiyono, K. Kuzushima, Y. Sugaya, S. Izuta, Y. Nishiyama, and T. Tsurumi. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudoh, A., M. Fujita, L. Zhang, N. Shirata, T. Daikoku, Y. Sugaya, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156-8163. [DOI] [PubMed] [Google Scholar]
- 33.Lew, D. J., and S. Kornbluth. 1996. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr. Opin. Cell Biol. 8:795-804. [DOI] [PubMed] [Google Scholar]
- 34.Lin, C. T., C. I. Wong, W. Y. Chan, K. W. Tzung, J. K. Ho, M. M. Hsu, and S. M. Chuang. 1990. Establishment and characterization of two nasopharyngeal carcinoma cell lines. Lab. Investig. 62:713-724. [PubMed] [Google Scholar]
- 35.Lou, Z., K. Minter-Dykhouse, and J. Chen. 2005. BRCA1 participates in DNA decatenation. Nat. Struct. Mol. Biol. 12:589-593. [DOI] [PubMed] [Google Scholar]
- 36.Lu, C. C., and M. R. Chen. 2006. Lytic replication of Epstein-Barr virus. Future Virol. 1:435-446. [Google Scholar]
- 37.Lu, C. C., Y. Y. Jeng, C. H. Tsai, M. Y. Liu, S. W. Yeh, T. Y. Hsu, and M. R. Chen. 2006. Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology 345:358-372. [DOI] [PubMed] [Google Scholar]
- 38.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maresca, T. J., and R. Heald. 2006. The long and the short of it: linker histone H1 is required for metaphase chromosome compaction. Cell Cycle 5:589-591. [DOI] [PubMed] [Google Scholar]
- 40.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 41.Mauser, A., E. Holley-Guthrie, A. Zanation, W. Yarborough, W. Kaufmann, A. Klingelhutz, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J. Virol. 76:12543-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 43.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 44.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 45.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20:214-220. [DOI] [PubMed] [Google Scholar]
- 46.Nurse, P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344:503-508. [DOI] [PubMed] [Google Scholar]
- 47.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shapiro, P. S., A. M. Whalen, N. S. Tolwinski, J. Wilsbacher, S. J. Froelich-Ammon, M. Garcia, N. Osheroff, and N. G. Ahn. 1999. Extracellular signal-regulated kinase activates topoisomerase IIα through a mechanism independent of phosphorylation. Mol. Cell. Biol. 19:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumner, A. T. 1995. Inhibitors of topoisomerase II delay progress through mitosis and induce a doubling of the DNA content in CHO cells. Exp. Cell Res. 217:440-447. [DOI] [PubMed] [Google Scholar]
- 54.Swenson, J. J., A. E. Mauser, W. K. Kaufmann, and S. C. Kenney. 1999. The Epstein-Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J. Virol. 73:6540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takada, K., K. Horinouchi, Y. Ono, T. Aya, T. Osato, M. Takahashi, and S. Hayasaka. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes 5:147-156. [DOI] [PubMed] [Google Scholar]
- 56.Takemoto, A., K. Kimura, S. Yokoyama, and F. Hanaoka. 2004. Cell cycle-dependent phosphorylation, nuclear localization, and activation of human condensin. J. Biol. Chem. 279:4551-4559. [DOI] [PubMed] [Google Scholar]
- 57.Tsurumi, T., M. Fujita, and A. Kudoh. 2005. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 15:3-15. [DOI] [PubMed] [Google Scholar]
- 58.Uemura, T., H. Ohkura, Y. Adachi, K. Morino, K. Shiozaki, and M. Yanagida. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50:917-925. [DOI] [PubMed] [Google Scholar]
- 59.Wang, J. T., P. W. Yang, C. P. Lee, C. H. Han, C. H. Tsai, and M. R. Chen. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 86:3215-3225. [DOI] [PubMed] [Google Scholar]
- 60.Yamashiro, S., and F. Matsumura. 1991. Mitosis-specific phosphorylation of caldesmon: possible molecular mechanism of cell rounding during mitosis. Bioessays 13:563-568. [DOI] [PubMed] [Google Scholar]
- 61.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 62.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 79:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.