Abstract
The 3A protein of coxsackievirus B3 (CVB3), a small membrane protein that forms homodimers, inhibits endoplasmic reticulum-to-Golgi complex transport. Recently, we described the underlying mechanism by showing that the CVB3 3A protein binds to and inhibits the function of GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factor 1 (Arf1), thereby interfering with Arf1-mediated COP-I recruitment. This study was undertaken to gain more insight into the molecular determinants underlying the interaction between 3A and GBF1. Here we show that 3A mutants that have lost the ability to dimerize are no longer able to bind to GBF1 and trap it on membranes. Moreover, we identify a conserved region in the N terminus of 3A that is crucial for GBF1 binding but not for 3A dimerization. Analysis of the binding domain in GBF1 showed that the extreme N terminus, the dimerization/cyclophilin binding domain, and the homology upstream of Sec7 domain are required for the interaction with 3A. In contrast to that of full-length GBF1, overexpression of a GBF1 mutant lacking its extreme N terminus failed to rescue the effects of 3A. Together, these data provide insight into the molecular requirements of the interaction between 3A and GBF1.
Enteroviruses (e.g., coxsackievirus, poliovirus, and echovirus) belong to the family Picornaviridae. They are nonenveloped, cytolytic viruses that contain a small positive-strand RNA genome. The viral RNA encodes a single large polyprotein that is processed into the individual capsid proteins and nonstructural proteins. The nonstructural proteins are involved in viral RNA replication and account for the virus-induced alterations in host cell metabolism and structure which serve to create an environment suitable for efficient viral RNA replication and/or to suppress antiviral host cell responses (24, 29, 34). Enteroviruses do not rely on an intact secretory pathway to release their virus progeny. Instead, they have been shown to induce a general blockage of protein secretion. Inhibition of protein secretion is also observed upon individual expression of the nonstructural proteins 2B and 3A (12, 30). The 3A-mediated inhibition of protein transport is not essential for virus replication but most likely serves to suppress antiviral host cell responses, such as cytokine secretion and antigen presentation (7, 11, 32).
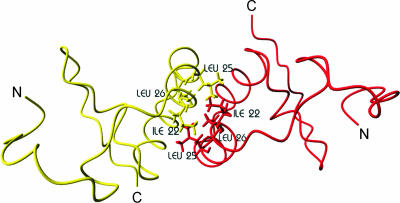
Coxsackievirus B3 (CVB3) 3A is a small (89 amino acids [aa]) integral membrane protein that is anchored in the membrane through its C-terminal hydrophobic domain (32). Elucidation of the structure of the soluble, cytosolic region upstream of the membrane anchor of the poliovirus 3A protein by nuclear magnetic resonance suggested that 3A forms homodimers (27). Each monomer was proposed to consist of two amphipathic α-helices, which are bent 180° to form a helical hairpin flanked by unstructured N and C termini. The structural data suggested that dimerization was mediated by hydrophobic surfaces, formed mainly by hydrophobic residues in the first amphipathic α-helix (Fig. 1). Based on this structure, we generated a molecular model of CVB3 3A and used this model for an extensive structure-function relationship study of CVB3 3A (31). Through this approach, experimental evidence of the functional relevance of CVB3 3A dimerization and important insight into the structural requirements for dimerization were obtained. The results supported the importance of a hydrophobic interaction between the monomers for dimerization. In addition, regions that are important for 3A functioning but not for dimerization were identified (31).
FIG. 1.
Molecular model of homodimer formed by N-terminal 60 aa of CVB3 3A. The construction of this molecular model is described in reference 31. N, N terminus; C, C terminus. Hydrophobic residues in the first α-helix that are implicated in dimerization are indicated.
Recently, we elucidated the mechanism by which the CVB3 3A protein inhibits protein transport (32). We showed that expression of 3A resulted in the disassembly of the vesicular tubular cluster and the Golgi complex. As a result, components of the vesicular tubular cluster and the Golgi complex flow back to the endoplasmic reticulum (ER) and accumulate at or close to the COP-II-coated ER exit sites, where 3A is also localized. 3A inhibits ER-to-Golgi complex transport by blocking activation of the ADP-ribosylation factor 1 (Arf1) protein. Arf proteins play a central role in protein transport and organelle structure and maintenance (15). Like other small GTPases, Arf proteins cycle between inactive GDP-bound and active GTP-bound states. Nucleotide exchange on Arfs is catalyzed by guanine nucleotide exchange factors (GEFs) (14). GBF1 is a GEF that has been implicated in ER-to-Golgi complex transport (4, 35). Under physiological conditions, GBF1 continuously cycles on and off membranes (23, 28). We showed that the CVB3 3A protein inhibits activation of Arf1 through interacting with and inhibiting the function of GBF1 (32). As a result, the COP-I coat complex, which plays an important role in bidirectional transport between the ER and the Golgi complex (26), cannot be recruited to membranes, and protein transport is inhibited. The poliovirus 3A protein was recently also shown to modify Arf1 membrane association (1).
This study was undertaken to gain more insight into the molecular determinants of the CVB3 3A-GBF1 interaction. To this end, we tested the ability of a number of 3A mutants to interfere with COP-I recruitment. Furthermore, the effects of 3A mutations on GBF1 binding and dynamics were investigated. Finally, we identified regions in the N terminus of GBF1 that are important for the interaction with 3A.
MATERIALS AND METHODS
Cells and viruses.
Buffalo green monkey (BGM) kidney cells were grown in minimal essential medium (Gibco) supplemented with 10% fetal bovine serum. COS-1 cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum. Cells were grown at 37°C in a 5% CO2 incubator. BGM cell monolayers were grown to subconfluence on coverslips in 24-well plates for immunofluorescence assays or in glass-bottomed dishes (WillCo Wells BV) for fluorescence recovery after photobleaching (FRAP) experiments and then transfected with 0.5 μg or 1 μg of plasmid DNA, respectively. Transfections were performed using FuGENE 6 (Roche) according to the manufacturer's instructions.
Plasmids. (i) p3A-myc constructs.
The plasmids coding for the mutant 3A-Myc proteins were generated by PCR amplification, using pGFP-3A mutants as templates. The forward and reverse primers for all mutants except the 3A-ins16S mutant introduced SalI and BamHI sites, respectively. The forward and reverse primers for the 3A-ins16S mutant introduced EcoRI and BamHI sites, respectively. The PCR products were cloned into p3A-myc (30), from which the 3A coding sequence was removed using the restriction enzymes SalI and BamHI. Sequence analysis showed that all PCR products contained correct sequences.
(ii) pCFP-3A constructs.
pCFP fusion constructs were generated by replacing the green fluorescent protein (GFP) coding sequence with the cyan fluorescent protein (CFP) coding sequence, using the restriction enzymes NheI and SspBI.
(iii) pGBT9-3A constructs.
pGBT9-3A was described before (32). The other constructs were generated by PCR amplifying the 3A coding sequences of the mutants and cloning the PCR products into pGBT9, using the restriction enzymes EcoRI and BamHI.
(iv) Mammalian two-hybrid constructs.
pACT-3A was described before (30). pACT-3A mutant plasmids were obtained by replacing the wild-type (wt) 3A sequence with mutant 3A sequences by using the enzymes Bst1107I and BamHI. Deletion mutants of GBF1 were first generated as yellow fluorescent protein (YFP) fusion proteins by PCR amplification. The PCR products were cloned into pEYFP-C1 (Clontech), using the restriction enzymes BglII and SalI. Subsequently, the GBF1 coding sequences were removed using the restriction enzymes BclI and SalI and cloned into pBIND that was cut with BamHI and KpnI.
(v) Venus-GBF1 constructs.
Venus is a variant of YFP (22) and is referred to as YFP in the text. YFP-GBF1 was described previously (23). YFP-GBF1ΔN was constructed by deleting nucleotides 1 to 111 from the GBF1 coding region.
Immunofluorescence.
Immunofluorescence assays were performed as described before (9). The effects of 3A mutant proteins on COP-I were studied by staining 3A-Myc-expressing cells with a monoclonal anti-Myc antiserum (diluted 1:20; Sigma) and a polyclonal anti-COP-I (against α- and γ-COP) antiserum (diluted 1:200) (from K. Frey and F. Wieland, Biochemie-Zentrum, Heidelberg, Germany). Pictures were taken under a Leica TCS NT microscope (Leica Lasertechnik GmbH, Heidelberg, Germany). Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin G and Alexa Fluor 488-conjugated anti-mouse immunoglobulin G were obtained from Molecular Probes.
FRAP experiments.
FRAP experiments were performed on a Zeiss LSM510Meta confocal microscope (Carl Zeiss GmbH, Jena, Germany) as described previously (32). Briefly, FRAP measurements were performed on time-lapse series that were taken at 37°C at a rate of 1 frame per s, using a 63×, 1.4-numerical-aperture objective and pinhole settings such that <-2 μm optical slices were imaged. Cells were selected on the basis of coexpression of CFP-3A and YFP-GBF1 or Arf1-YFP. Regions of interest were selectively photobleached, using the 514-nm line at 100% transmission.
Yeast two-hybrid analysis.
Yeast two-hybrid analysis was performed with strain AH109 (Clontech) as described previously (32). Briefly, the pGADT7 and pGBT9 fusion constructs were transformed into yeast by the lithium acetate method using carrier DNA and plated on nonselective plates (lacking leucine and tryptophan). After growth, the colonies were transferred to selective plates (lacking histidine or adenine).
Mammalian two-hybrid analysis.
COS cells grown in 24-well plates were transfected with a total of 0.75 μg plasmid DNA (1:1:1 mix of the pACT, pBIND, and pG5luc plasmids). At 48 h posttransfection, the cells were lysed, and both the firefly luciferase and Renilla luciferase enzyme activities were measured from the same cell lysate by use of a dual-luciferase reporter assay system (Promega) as described previously (8). An analysis of the Renilla luciferase activities, encoded by the pBIND plasmid and allowing monitoring of the transfection efficiency, revealed no gross differences in efficiencies of transfection among the different samples. All pACT- and pBIND-encoded fusion proteins were efficiently expressed (data not shown). The 3A-GBF1 interaction was expressed as the firefly luciferase activity in cells coexpressing 3A and GBF1 fusion proteins and was normalized to 100% for cells coexpressing wt 3A and the complete N terminus of GBF1. The firefly luciferase activity measured in cells coexpressing mutant 3A and GBF1 proteins was normalized to the activity measured in cells coexpressing wt 3A and GBF1 fusion proteins (which was set at 100% in each experiment).
Coimmunoprecipitation.
Coimmunoprecipitation experiments were performed as described previously (32). Briefly, GFP or YFP fusion proteins that were coexpressed with 3A-Myc in BGM cells were immunoprecipitated using an anti-GFP antibody (raised against recombinant glutathione S-transferase-GFP). The presence of 3A-Myc in these samples was checked by Western analysis using a polyclonal anti-Myc antiserum (Affinity BioReagents).
Statistical analysis.
In all graphs, data are expressed as means ± standard errors of the means. Overall statistical analysis was determined by analysis of variance followed by Scheffe's test to investigate significance between individual values and control values, using Origin Pro 7.5 software (Originlabs, Northampton, MA). P values below 0.05 were considered significant.
RESULTS AND DISCUSSION
Inhibition of COP-I recruitment by mutant 3A proteins.
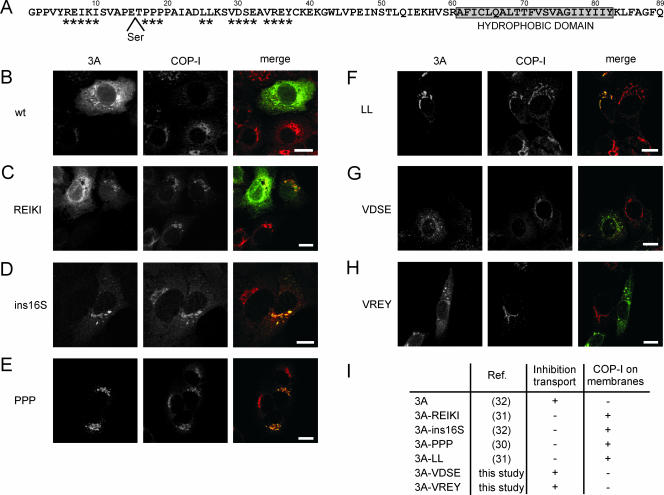
Previously, we constructed several 3A mutants and characterized them for the ability to dimerize and inhibit secretion of a reporter protein (30, 31, 32). A number of 3A mutants were obtained that were no longer able to inhibit reporter protein secretion. We reasoned that this might be due to an impaired ability to interfere with COP-I recruitment to membranes. To investigate this, C-terminal Myc fusions of a number of selected 3A mutants were generated. These mutants are summarized in Fig. 2A and described below (in order of the positions of the mutations, from the N to the C terminus). The addition of a C-terminal Myc tag did not interfere with the abilities of 3A to dimerize (data not shown) and to inhibit protein transport (13, 32).
FIG. 2.
Inhibition of COP-I recruitment to membranes by mutant 3A proteins. (A) Amino acid sequence of CVB3 3A. The C-terminal hydrophobic domain (aa 61 to 82) is depicted in the boxed area. Amino acids that are mutated are indicated by asterisks, and the Ser insertion at position 16 is also indicated. (B to H) BGM cells expressing Myc-tagged wt 3A (B), 3A-REIKI (C), 3A-ins16S (D), 3A-PPP (E), 3A-LL (F), 3A-VDSE (G), and 3A-VREY (H) were stained for the Myc tag and COP-I. (I) Table summarizing the ability of 3A mutants to inhibit protein transport and COP-I recruitment. References in which the mutants are described are indicated. 3A-REIKI, 3A-R6A/E7A/I8A/K9A/I10A; 3A-PPP, 3A-P17A/P18A/P19A; 3A-LL, 3A-L25A/L26A; 3A-VDSE, 3A-V29A/D30A/S31A/E32A; 3A-VREY, 3A-V34A/R35A/E36A/Y37A. Bars, 10 μm.
(i) 3A-R6A/E7A/I8A/K9A/I10A is a mutant in which Arg6, Glu7, Ile8, Lys9, and Ile10 are replaced with Ala residues. These residues are located in the N terminus of 3A, a region that was predicted to be unstructured and not to be involved in dimerization. Indeed, we found that mutant 3A-R6A/E7A/I8A/K9A/I10A showed efficient dimerization. Nevertheless, this mutant was unable to inhibit reporter protein secretion. (ii) 3A-ins16S is a mutant in which a Ser residue is inserted at position 16 in 3A. (iii) 3A-P17A/P18A/P19A is a mutant in which Pro17, Pro18, and Pro19 are replaced with Ala residues. The last two mutants contain amino acid alterations in the region immediately upstream of the first α-helix (aa 20 to 27). Although this region was not predicted to be important for dimerization, both mutants were defective in 3A dimerization and inhibition of reporter protein secretion, which may be due to overall effects on protein folding. (iv) 3A-L25A/L26A is a mutant in which Leu25 and Leu26 are replaced with Ala residues. These residues are located in the first α-helix and predicted to be involved in the hydrophobic packing between the 3A monomers. Consistent with this, mutant 3A-L25A/L26A was unable to dimerize and inhibit secretion of a reporter protein. For reasons of simplicity, mutants 3A-R6A/E7A/I8A/K9A/I10A, 3A-P17A/P18A/P19A, and 3A-L25A/L26A are referred to in this study as 3A-REIKI, 3A-PPP, and 3A-LL, respectively.
We also generated C-terminal Myc fusions of two new 3A mutants. These mutants contain substitutions of residues in the middle part of 3A that are conserved among the different enteroviruses. The mutants are (v) 3A-VDSE, a mutant in which Val29, Asp30, Ser31, and Glu32 are replaced with Ala residues, and (vi) 3A-VREY, a mutant in which Val34, Arg35, Glu36, and Tyr37 are replaced with Ala residues. Both mutants were able to inhibit reporter protein secretion and formed dimers efficiently in mammalian two-hybrid assays (data not shown).
Immunofluorescence microscopy of cells, using anti-COP-I antibodies, showed that COP-I was typically localized at Golgi membranes in control cells. In line with previous results, COP-I was redistributed to the cytoplasm in cells expressing the wt 3A protein (Fig. 2B) (32). In these cells, 3A localized at the ER and at or close to punctate structures (Fig. 2B), which we have demonstrated previously to represent rearranged membranes containing markers for COP-II, ER-Golgi intermediate compartment, and the Golgi complex (32). In cells expressing 3A-REIKI, 3A-ins16S, 3A-PPP, and 3A-LL (i.e., the 3A mutants that were unable to inhibit protein transport), COP-I was localized at Golgi membranes (Fig. 2C to F and I). The 3A-ins16S, 3A-PPP, and 3A-LL mutants were found to localize at the Golgi complex (Fig. 2D, E, and F), whereas the 3A-REIKI mutant localized mainly to the ER and partially to the Golgi complex (Fig. 2C). In contrast, the 3A-VDSE and 3A-VREY mutants (i.e., the 3A mutants that were able to inhibit protein transport) localized similarly to wt 3A and redistributed COP-I to the cytoplasm (Fig. 2G, H, and I). Together, the data demonstrate that defective 3A mutants cannot inhibit protein transport since they can no longer interfere with COP-I recruitment.
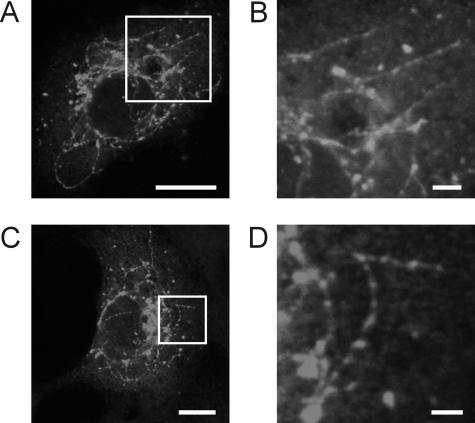
Interestingly, long thread-like tubulovesicular structures could be observed in cells expressing wt 3A (Fig. 3) or mutant 3A proteins that were still able to inhibit transport. These structures contained 3A, an integral membrane protein, at their surfaces and were therefore most likely membrane tubules. No such structures were observed in cells expressing any of the defective 3A mutants.
FIG. 3.
Membrane tubules in 3A-expressing BGM cells. (A and C) BGM cells expressing Myc-tagged wt 3A, stained for the Myc tag. (B and D) Higher-magnification pictures of the parts of the cells indicated by the white boxes in panels A (B) and C (D), showing the tubules in more detail. Bars, 10 μm (A and C) and 2 μm (B and D).
Ability of mutant 3A proteins to bind GBF1 and trap it on membranes.
GBF1 is an Arf1 GEF that normally cycles rapidly on and off Golgi membranes (23, 28). GBF1-mediated activation of Arf1 is required for COP-I recruitment to membranes. We showed that wt 3A interferes with COP-I recruitment by binding to (the N-terminal part of) GBF1 and trapping it on membranes (32). The observation that the 3A mutants described above are defective in interfering with COP-I recruitment may be explained by their inability to bind GBF1. Alternatively, these mutants may still bind GBF1 but no longer be able to trap it on membranes. To discriminate between these possibilities, binding of these mutants to GBF1, as well as their effects on GBF1 dynamics, was tested.
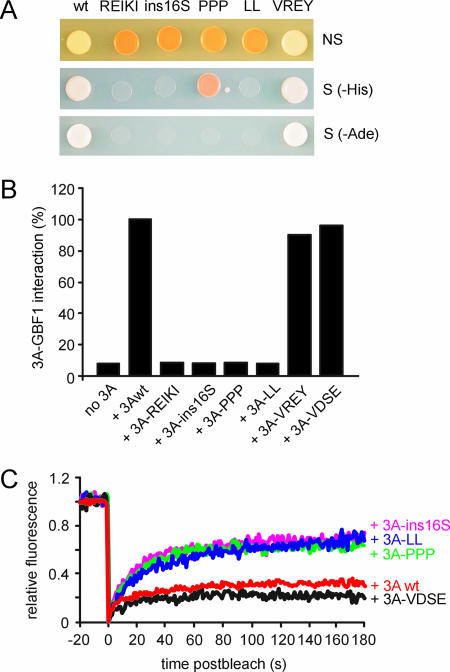
Binding of the 3A mutants to the N-terminal part of GBF1 was investigated by both yeast two-hybrid (Fig. 4A) and mammalian two-hybrid (Fig. 4B) analyses. In Saccharomyces cerevisiae, the cytosolic part of 3A (i.e., the 60 aa upstream of its hydrophobic membrane anchor) was expressed, whereas full-length 3A was expressed in the mammalian system. All 3A mutants that were unable to inhibit protein transport and to inhibit COP-I recruitment to membranes (3A-REIKI, 3A-ins16S, 3A-PPP, and 3A-LL) were severely reduced in the ability to bind GBF1 (Fig. 4A and B). In contrast, the two mutants that were able to inhibit protein transport and to cause release of COP-I into the cytoplasm (3A-VDSE and 3A-VREY) showed a strong interaction with GBF1 (Fig. 4A and B). These data clearly indicate that the 3A-REIKI, 3A-ins16S, 3A-PPP, and 3A-LL mutants are unable to inhibit COP-I recruitment and protein transport because they cannot bind GBF1.
FIG. 4.
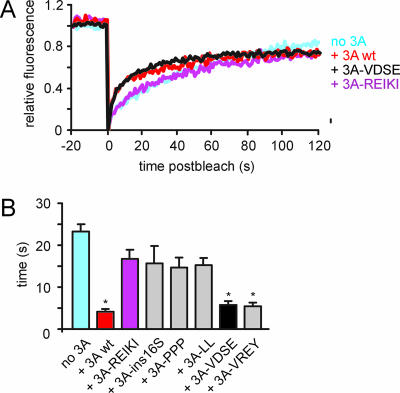
Interaction of 3A mutants with GBF1 and effects on GBF1 dynamics. (A) Interaction of wt 3A, 3A-REIKI, 3A-ins16S, 3A-PPP, 3A-LL, and 3A-VREY with the N-terminal part of GBF1 in yeast two-hybrid analysis. The upper, middle, and lower panels show growth of yeast on nonselective medium (leucine- and tryptophan-deficient medium [NS]), selective medium lacking histidine (−His), and selective medium lacking adenine (−Ade), respectively. (B) Interaction of wt 3A, 3A-REIKI, 3A-ins16S, 3A-PPP, 3A-LL, and 3A-VREY with the N-terminal part of GBF1 in mammalian two-hybrid analysis. The firefly luciferase activities measured at 48 h posttransfection are depicted. The activity measured with wt 3A and the GBF1 N terminus was set at 100%. (C) Dynamics of YFP-GBF1. FRAP traces were done for cells expressing YFP-GBF1 together with CFP fusion proteins of wt 3A, 3A-ins16S, 3A-PPP, 3A-LL, and 3A-VDSE. The traces show the average recoveries (n ≥ 10 cells) for at least two independent experiments. The fluorescence intensity before bleaching was normalized to 1, and the fluorescence intensity directly after bleaching was normalized to 0. The fluorescence intensity was corrected for bleaching of the cell during imaging and for background fluorescence.
Putative effects of the 3A mutant proteins on the dynamics of GBF1 were tested using FRAP. FRAP measurements were performed on dispersed membrane structures that contained YFP-GBF1 and CFP-3A (32). We compared GBF1 dynamics in cells expressing wt 3A, 3A-ins16S, 3A-PPP, 3A-LL, and 3A-VDSE. GBF1 dynamics in cells expressing 3A-REIKI and 3A-VREY was not analyzed since in these cells, for reasons that are yet unknown, YFP-GBF1 localized at the ER rather than on dispersed membranes (which would not allow a proper comparison). The results showed that the 3A mutants that were unable to inhibit protein transport (3A-ins16S, 3A-PPP, and 3A-LL) were unable to trap GBF1 on membranes, whereas 3A-VDSE trapped GBF1 on membranes as efficiently as wt 3A (Fig. 4C). In summary, these findings indicate that 3A mutants that are unable to inhibit COP-I recruitment to membranes and to block protein transport are also unable to trap GBF1 on membranes.
Previously, we showed that expression of 3A resulted in an increased cycling time of Arf1-YFP (32). Although the underlying reason for this phenomenon is unclear, it seems likely that it is linked to the 3A-mediated inhibition of GBF1. To gain more insight into this possible correlation, we tested the effects of the 3A mutants on Arf1 dynamics (Fig. 5). In cells expressing 3A mutants that were able to inhibit GBF1 activity (3A-VDSE and 3A-VREY), the half-time of fluorescence recovery of Arf1-YFP was decreased to an extent similar to that for cells expressing wt 3A, whereas in cells expressing 3A mutants that were not able to inhibit protein transport (3A-REIKI, 3A-ins16S, 3A-PPP, and 3A-LL), the half-time of fluorescence recovery of Arf1-YFP was slightly reduced, though not significantly, compared to that for control cells (Fig. 5). These results provide support for a correlation between the activity of GBF1 and the dynamics of Arf1.
FIG. 5.
Effects of 3A mutants on Arf1 dynamics. The upper panel shows FRAP traces for cells expressing Arf1-YFP either alone (no 3A) or together with CFP fusion proteins of wt 3A, 3A-REIKI, and 3A-VDSE. The FRAP traces for Arf1-YFP in cells expressing 3A-ins16S, 3A-PPP, and 3A-LL were similar to that observed for cells expressing 3A-REIKI (and are therefore not shown). The FRAP trace for Arf1-YFP in cells expressing 3A-VREY was similar to that for cells expressing 3A-VDSE. The lower panel shows half-times of fluorescence recovery of Arf1-YFP expressed alone or together with wt 3A, 3A-REIKI, 3A-ins16S, 3A-PPP, 3A-LL, 3A-VDSE, or 3A-VREY. The traces show the average recoveries (n ≥ 10 cells) for at least two independent experiments. The fluorescence intensity was calculated as described in the legend to Fig. 4.
Identification of regions in the N terminus of GBF1 that are important for interaction with 3A.
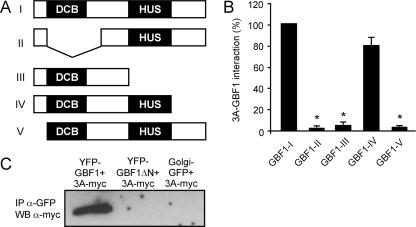
We also sought to identify the binding domain of 3A in GBF1. All ArfGEFs identified to date have a Sec7 domain, a conserved module of ∼200 aa that carries the guanine nucleotide exchange activity and that has been studied extensively (3, 16). In contrast to the Sec7 domain, only a little is known about the functions of the other GEF domains, which are likely to determine intracellular localization and intermolecular interactions (3, 6, 10, 18, 20). The N-terminal part of GBF1 has been shown to be sufficient for 3A binding (32). To gain more insight into the region(s) of GBF1 that is responsible for the interaction with 3A, we made deletion constructs of the N terminus of GBF1 (Fig. 6A) and tested them for interaction with 3A by mammalian two-hybrid analysis (Fig. 6B). Two homology domains have been identified in the N-terminal part of GBF1, namely, the dimerization/cyclophilin binding (DCB) domain and the homology upstream of Sec7 (HUS) domain (21). Deletion of the DCB domain (GBF1-II) completely abrogated the interaction with 3A. Combined deletion of the HUS domain and the C-terminal region downstream of this domain (GBF1-III) also abolished the interaction with 3A, whereas deletion of the C-terminal region alone (GBF1-IV) had no effect. Deleting the extreme N terminus, i.e., the region upstream of the DCB domain (GBF1-V), also abrogated the interaction with 3A. The importance of the latter region for the interaction with 3A was confirmed by showing that a GBF1 mutant in which the extreme N terminus was deleted from (full-length) GBF1 (GBF1ΔN) was no longer able to interact with 3A in coimmunoprecipitation experiments (Fig. 6C). Together, these data indicate that both the extreme N terminus of GBF1 and the DCB and HUS domains are important for the 3A-GBF1 interaction. It remains to be established whether each of these regions contains contact sites for the interaction with 3A or, alternatively, if different parts of the protein fold together into a higher-order structure that provides a binding site for 3A. Elucidation of the structure of GBF1 would help to further define the binding domain of 3A in GBF1.
FIG. 6.
Characterization of the interaction between GBF1 and 3A. (A) Schematic representation of deletion mutants of the N terminus of GBF1. (B) Interaction of 3A with GBF1 deletion mutants in a mammalian two-hybrid system. The firefly luciferase activities measured at 48 h posttransfection are depicted. Average values and standard errors of the means for three experiments are shown. In each experiment, the activity measured with 3A and the intact GBF1 N terminus was set at 100%. (C) Coimmunoprecipitation experiments. 3A-Myc was specifically coprecipitated (IP) from cells cotransfected with YFP-GBF1 but not from cells cotransfected with YFP-GBF1ΔN or Golgi complex-GFP. GBF1-I, aa 1 to 710 of GBF1; GBF1-II, aa 1 to 52 and 295 to 710 of GBF1; GBF 1-III, aa 1 to 392 of GBF1; GBF1-IV, aa 1 to 566 of GBF1; GBF1-V, aa 52 to 710 of GBF1; GBF1ΔN, GBF1 lacking aa 1 to 37. WB, Western blot.
A GBF1 mutant that lacks the extreme N terminus cannot rescue the effects of 3A.
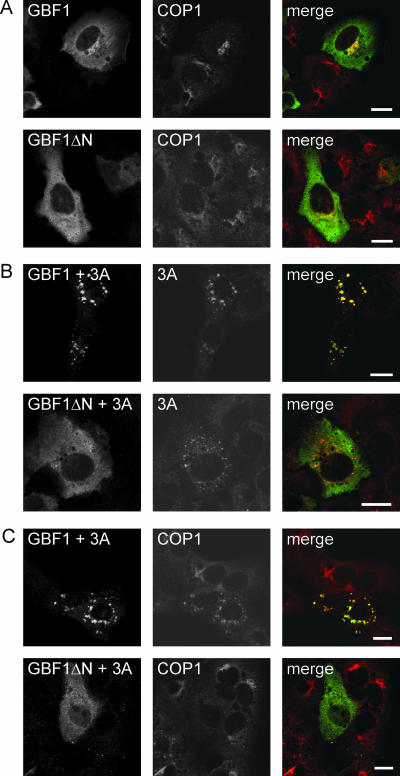
Previously, we showed that overexpression of GBF1 suppressed 3A function (32). Whereas COP-I is redistributed to the cytoplasm in almost all 3A-expressing cells, COP-I was still associated with membranes in most of the cells coexpressing GBF1 and 3A. In these cells, either an intact Golgi complex or dispersed membrane structures which contained 3A, GBF1, and several Golgi complex markers were observed (32). We sought to investigate whether the extreme N terminus of GBF1, which is important for the 3A-GBF1 interaction, is essential for this effect. Overexpression of GBF1 or GBF1ΔN had little, if any, effect on the localization of COP-I (Fig. 7A). Consistent with previous results, coexpression of GBF1 and 3A gave rise to dispersed membrane structures that contained both proteins and that also contained COP-I (Fig. 7B and C) (32). In contrast, COP-I was redistributed to the cytosol in cells coexpressing GBF1ΔN and 3A, indicating that the N terminus of GBF1 is required for its ability to counteract the effects of 3A (Fig. 7C). In these cells, 3A localization was similar to that in cells expressing 3A alone, and no colocalization with GBF1ΔN was observed (Fig. 7B). Thus, the extreme N terminus of GBF1 is required, not only for the interaction with 3A but also for the rescue of the 3A-mediated inhibition of Arf1 activation. Despite this correlation, it remains to be established whether an interaction between 3A and GBF1 is important for the rescue effect.
FIG. 7.
A GBF1 mutant that lacks the extreme N terminus cannot rescue the effects of 3A. (A) Cells expressing YFP-GBF1 or YFP-GBF1ΔN were stained for COP-I. (B) Images showing GBF1 (left) and 3A (middle). Cells expressing YFP-GBF1 and 3A-Myc or YFP-GBF1ΔN and 3A-Myc were stained for the Myc tag. (C) Images showing GBF1 (left) and COP-I (middle). Cells expressing YFP-GBF1 and 3A-Myc or YFP-GBF1ΔN and 3A-Myc were stained for COP-I. GBF1ΔN, GBF1 lacking aa 1 to 37. Bars, 10 μm.
Concluding remarks.
This study was undertaken to gain more insight into the molecular determinants that are required for the interaction between 3A and GBF1. We characterized a number of 3A mutants and found that mutants that were unable to inhibit protein transport were no longer able to inhibit COP-I recruitment because they were unable to bind to GBF1 and trap it on membranes. Previously, we showed that dimerization is important for efficient inhibition of protein transport (31). Here we showed that mutants that are impaired in dimerization, i.e., mutants 3A-ins16S, 3A-PPP, and 3A-LL (31, 32), are unable to bind GBF1, providing an explanation for their inability to inhibit transport. The 3A-REIKI mutant was also unable to inhibit protein transport. However, this mutant showed efficient dimerization (31), and therefore we speculated that the REIKI region might be involved in binding of a cellular partner. Here we provided experimental evidence for this hypothesis by showing that the 3A-REIKI mutant is no longer able to bind GBF1. We hypothesize that dimerization is required for efficient exposure of the REIKI residues. In addition, dimerization may create other contact sites in 3A (i.e., other than the REIKI region) for the interaction with GBsF1.
The wt 3A protein localizes at the ER and a dispersed post-ER compartment, but most 3A mutants that were defective in inhibiting protein transport localized at the Golgi complex. It can be speculated that upon its generation, 3A is first transported to the Golgi complex, where the majority of endogenous GBF1 is localized (5, 17, 35). Inhibition of GBF1 by 3A will lead to inactivation of Arf1, resulting in dissociation of COP-I from Golgi membranes. This will lead to disassembly of the Golgi complex since COP-I is required for maintenance of the structural integrity of the Golgi complex (19). Treatment of cells with brefeldin A has been shown to result in rapid Golgi complex disassembly and in redistribution of the Golgi proteins into the ER via membrane tubules that are formed in a highly dynamic manner and that mediate retrograde transport to the ER (19, 25). Membrane tubules were also observed in 3A-expressing cells. Thus, as in brefeldin A-treated cells, Golgi complex disassembly and backflow into the ER in 3A-expressing cells probably occur via membrane tubulation. 3A mutants that cannot inhibit GBF1 function and COP-I recruitment will remain localized at the Golgi complex.
Recently, we showed that not only the 3A protein of CVB3 but also that of the closely related poliovirus (PV) inhibits ER-to-Golgi complex transport by binding and inhibiting GBF1 (33). While this work was in progress, Belov et al. reported that Arf1 is recruited to membranes in PV-infected cells and that Arf1-GTP levels increase approximately fourfold during infection (2). The reason for the difference between their data (i.e., recruitment and increased activation of Arf1 during infection in infected cells) and our data (i.e., inactivation of Arf1 and redistribution to the cytosol during expression of 3A alone) is still unknown. A possible explanation is that more (viral) factors modify the activation state of Arf1 during infection. Indeed, Belov et al. showed that in addition to the PV 3A protein, which was found to interact with GBF1, the PV 3CD protein also interacts with ArfGEFs (i.e., BIG1 and BIG2). Thus, the regulation of Arf1 in infected cells seems to be modulated by multiple viral proteins in a complex, and still poorly understood, way.
Acknowledgments
We thank Jack Fransen, Huib Croes (Microscopical Imaging Centre Nijmegen), and Henri Dijkman for assistance with microscopy and Werner Koopman for help with statistical analysis.
This work was partly supported by grants from The Netherlands Organization for Scientific Research (NWO-VIDI-917.46.305) and the M. W. Beijerink Virology Fund from the Royal Netherlands Academy of Sciences.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Belov, G. A., M. H. Fogg, and E. Ehrenfeld. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 79:7207-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belov, G. A., N. Altan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chardin, P., S. Paris, B. Antonny, S. Robineau, S. Beraud-Dufour, C. L. Jackson, and M. Chabre. 1996. A human exchange factor for Arf contains Sec7- and pleckstrin-homology domains. Nature 384:481-484. [DOI] [PubMed] [Google Scholar]
- 4.Claude, A., B. P. Zhao, C. E. Kuziemsky, S. Dahan, S. J. Berger, J. P. Yan, A. D. Armold, E. M. Sullivan, and P. Melancon. 1999. GBF1: a novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146:71-84. [PMC free article] [PubMed] [Google Scholar]
- 5.Cornell, C. T., W. B. Kiosses, S. Harkins, and J. L. Whitton. 2006. Inhibition of protein trafficking by coxsackievirus B3: multiple viral proteins target a single organelle. J. Virol. 80:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen, P. J., and P. Chardin. 2000. Membrane targeting: what a difference a G makes. Curr. Biol. 10:R876-R878. [DOI] [PubMed] [Google Scholar]
- 7.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong, A. S., I. W. Schrama, P. H. Willems, J. M. Galama, W. J. Melchers, and F. J. van Kuppeveld. 2002. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83:783-793. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, A. S., E. Wessels, H. B. P. M. Dijkman, J. M. D. Galama, W. J. G. Melchers, P. H. M. G. Willems, and F. J. M. van Kuppeveld. 2003. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 278:1012-1021. [DOI] [PubMed] [Google Scholar]
- 10.Derrien, V., C. Couillault, M. Franco, S. Martineau, P. Montcourrier, R. Houlgatte, and P. Chavrier. 2002. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 115:2867-2879. [DOI] [PubMed] [Google Scholar]
- 11.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, J. G., and C. L. Jackson. 2000. Regulators and effectors of the Arf GTPases. Curr. Opin. Cell Biol. 12:475-482. [DOI] [PubMed] [Google Scholar]
- 15.D'Souza-Schorey, C., and P. Chavrier. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 7:347-358. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, C. L., and J. E. Casanova. 2000. Turning on Arf: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10:60-67. [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto, K., Y. Yoshida, H. Tamaki, S. Torii, C. Shinotsuka, S. Yamashina, and K. Nakayama. 2002. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 7:483-495. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. Y., and B. Pohajdak. 2000. N-terminal targeting of guanine nucleotide exchange factors (GEF) for ADP ribosylation factors (ARF) to the Golgi. J. Cell Sci. 113:1883-1889. [DOI] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz, J., J. G. Donaldson, A. Schweizer, E. G. Berger, H.-P. Hauri, L. C. Yuan, and R. D. Klausner. 1990. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell 60:821-836. [DOI] [PubMed] [Google Scholar]
- 20.Mansour, M., S. Y. Lee, and B. Pohajdak. 2002. The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J. Biol. Chem. 277:32302-32309. [DOI] [PubMed] [Google Scholar]
- 21.Mouratou, B., V. Biou, A. Joubert, J. Cohen, D. J. Shields, N. Geldner, G. Jürgens, P. Melancon, and J. Cherfils. 2005. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90.11753368 [Google Scholar]
- 23.Niu, T.-K., A. C. Pfeifer, J. Lippincott-Schwartz, and C. L. Jackson. 2005. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell 16:1213-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter, A. G. 1993. Picornavirus nonstructural proteins: emerging roles in virus replication and inhibition of host cell functions. J. Virol. 67:6917-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Presley, J. F., C. Smith, K. Hirschberg, C. Miller, N. B. Cole, K. J. M. Zaal, and J. Lippincott-Schwartz. 1998. Golgi membrane dynamics. Mol. Biol. Cell 9:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabouille, C., and J. Klumperman. 2005. The maturing role of COPI vesicles in intra-Golgi transport. Nat. Rev. Mol. Cell. Biol. 6:812-817. [DOI] [PubMed] [Google Scholar]
- 27.Strauss, D. M., L. W. Glustrom, and D. S. Wuttke. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330:225-234. [DOI] [PubMed] [Google Scholar]
- 28.Szul, T., R. Garcia-Mata, E. Brandon, S. Shestopal, C. Alvarez, and E. Sztul. 2005. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6:374-385. [DOI] [PubMed] [Google Scholar]
- 29.van Kuppeveld, F. J., A. S. de Jong, W. J. Melchers, and P. H. Willems. 2005. Enterovirus protein 2B po(u)res out the calcium: a viral strategy to survive. Trends Microbiol. 13:41-44. [DOI] [PubMed] [Google Scholar]
- 30.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. Melchers, and F. J. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79:5163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wessels, E., R. Notebaart, D. Duijsings, K. Lanke, B. Vergeer, W. Melchers, and F. van Kuppeveld. 2006. Structure-function analysis of the coxsackievirus protein 3A: identification of residues important for dimerization, viral RNA replication, and transport inhibition. J. Biol. Chem. 281:28232-28243. [DOI] [PubMed] [Google Scholar]
- 32.Wessels, E., D. Duijsings, T. Niu, S. Neumann, V. Oorschot, F. de Lange, K. H. Lanke, J. Klumperman, A. Henke, C. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell 11:191-201. [DOI] [PubMed] [Google Scholar]
- 33.Wessels, E., D. Duijsings, K. H. Lanke, S. H. van Dooren, C. L. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J. Virol. 80:11852-11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, X., T. K. Lasell, and P. Melancon. 2002. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi complex compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell 13:119-133. [DOI] [PMC free article] [PubMed] [Google Scholar]