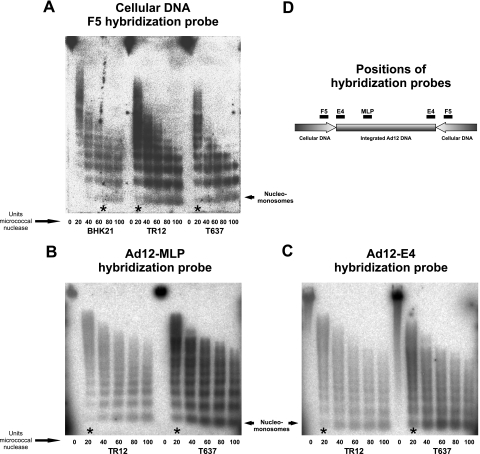
FIG. 5.
Chromatin organization in Ad12 transgenomes and their vicinity. The degree of chromatin condensation was determined by assessing the susceptibility to micrococcal nuclease (see Materials and Methods) (A) in the F5 cellular DNA segments of BHK21, T637, and TR12 cells and in the Ad12 region MLP (B) and E4 promoter (C) in cell lines T637 and TR12. Micrococcal nuclease units sufficient to generate nucleomonosomes were marked with asterisks. (D) Map positions of hybridization probes. As the amounts of Ad12 DNA integrates present in cell lines T637 and TR12 differ by a factor of about 15, signal intensities are expected to be different when Ad12 hybridization probes are used. The data in Fig. 5A demonstrate that adequate amounts of TR12 DNA had been employed in the experiments shown in this figure. In the panel A experiments, an F5 hybridization probe from hamster cellular DNA was used. TR12 and T637 DNAs contain the same equivalents of cellular DNA segments, like F5, but differ by a factor of about 15 in the content of Ad12 DNA sequences. Therefore, the blots of TR12 DNA are expected to exhibit weaker Ad12 DNA signals (B and C).