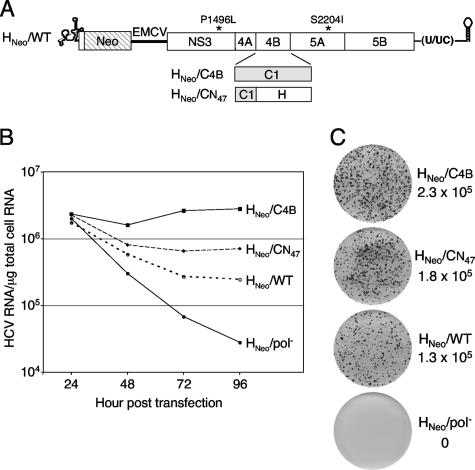
FIG. 5.
HCV RNA accumulation and G418-resistant colony-forming abilities of H77-derived subgenomic replicons. (A) Structure of the G418-selectable replicon. The 5′ and 3′ NTR structures are shown, and the H77 NS3-5B ORF is depicted as an open box; the polyprotein cleavage products are indicated, along with the positions of the two cell culture-adaptive mutations, P1496L and S2204I. The neomycin phosphotransferase gene (Neo) and the encephalomyocarditis virus (EMCV) IRES are shown. Chimeric NS4B proteins are diagramed below, with the shaded regions representing Con1-derived NS4B sequences and the adopted nomenclature given on the left. (B) Total cellular RNA was harvested at the indicated times postelectroporation, and HCV RNA in 1 μg of total RNA was quantified by real-time RT-PCR. (C) G418 transduction efficiencies of H77 replicons. Immediately after electroporation, 1,250, 2,500, 5,000, 104, or 2.5 × 104 transfected Huh-7.5 cells were coplated with cells transfected with HNeo/pol− RNA as described in Materials and Methods. Following 3 weeks of G418 selection, the resulting colonies were stained with crystal violet and counted for at least three cell densities. The calculated transduction efficiencies (expressed in CFU per μg of transfected RNA) are given to the right of representative dishes that were initially plated with 2.5 × 104 transfected cells. Similar results were obtained in three independent repetitions of the experiment.