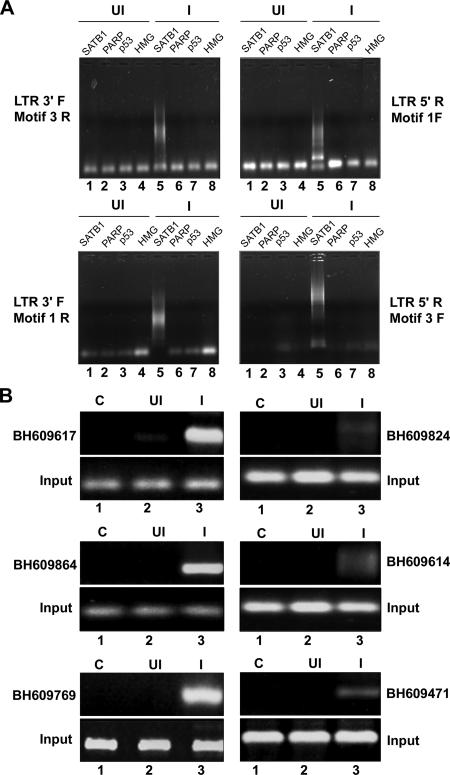
FIG. 2.
ChIP-PCR analysis of DNA flanking integration sites from the pool of SATB1-bound DNA. (A) ChIP analysis of regions flanking HIV-1 integration sites. In vivo association of SATB1 with the regions flanking integration sites was monitored by ChIP-PCR as described in Materials and Methods. DNA fragments isolated from infected (I) and control (uninfected [UI]) cell chromatin after immunoprecipitation with antibodies against four chromatin proteins [SATB1, PARP, p53, and HMG-I(Y)] were used as templates for PCR amplification with primer sets containing an LTR-specific primer and a primer specific for an Alu-like motif. The combinations of primers used are indicated at the sides. (B) PCR amplification of reported integration sequences (BH series) in the SATB1-immunoprecipitated and PCR-amplified DNA pool with the LTR 3′F and Motif 3R primers. We selected the indicated clones that bind SATB1 in vitro and confirmed in vivo association with SATB1 by PCR with primer sets corresponding to each of them and ChIP-PCR-amplified DNA as the template.