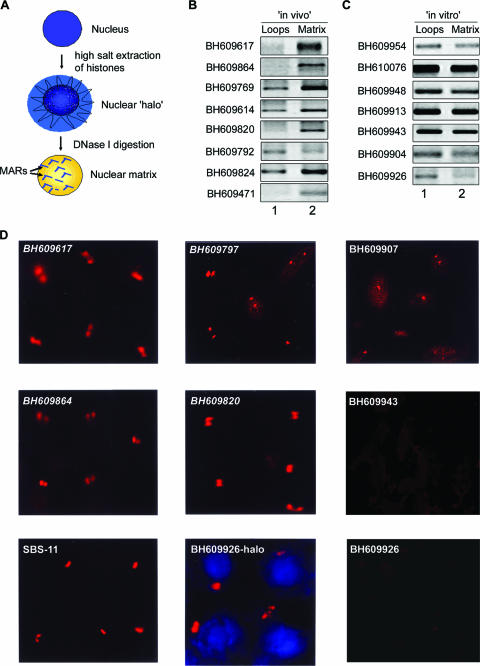
FIG. 3.
Regions flanking HIV-1 integration sites partition preferentially in the chromatin fraction associated with the nuclear matrix. (A) Schematic representation of the protocol used for the preparation of nuclear halos and matrices. For details, see Materials and Methods. (B and C) The matrix-loop partitioning assay was performed as described in Materials and Methods. PCR amplification of the indicated in vivo (B) or in vitro (C) integration sequences with template DNA from the loop (lane 1) or matrix (lane 2) fraction is depicted. The PCR products were resolved on 1% agarose and visualized by staining with ethidium bromide. (D) Amplified single-locus FISH revealed that cloned sequences flanking in vivo HIV-1 integration sites are associated specifically with the nuclear matrix in situ. Briefly, nuclear matrices were prepared from Jurkat T cells in situ and PCR-amplified and biotin-labeled DNA probes corresponding to the integration clones (BH series) were used for hybridization and detected by amplified FISH as previously described (11). The in vivo integration clones are in italics. As a positive control for matrix hybridization, we used the reported SATB1-binding sequence SBS-11 (15) as depicted at the bottom left. BH609926-halo represents hybridization of the biotin-labeled probe corresponding to this in vitro integration clone to the nuclear halo comprising distended chromatin loops. The same probe did not hybridize with the nuclear matrix preparation, as depicted at the bottom right.