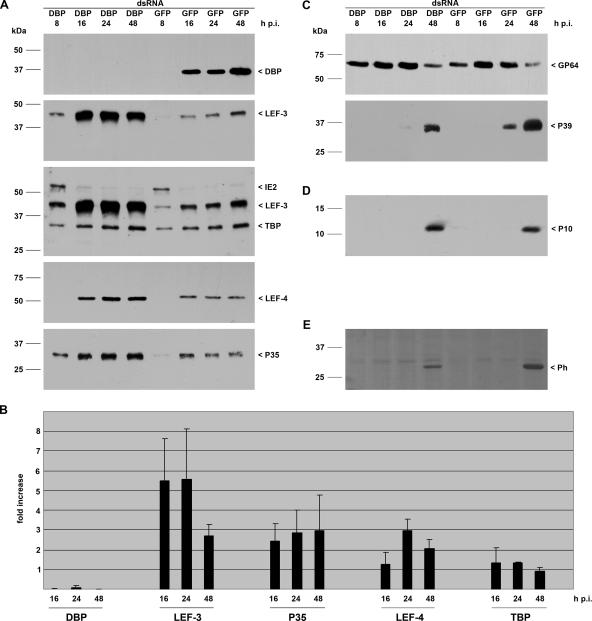
FIG. 1.
Effect of dbp silencing on viral gene expression. S. frugiperda cells were transfected with either DBP dsRNA or GFP dsRNA. Cells were subsequently infected with AcMNPV (10 PFU/cell) at 20 h posttransfection, and detergent-based nuclear and cytoplasmic extracts were prepared from infected cells 8, 16, 24, and 48 h p.i. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to nitrocellulose (A, C, and E) or resolved on sodium dodecyl sulfate-15% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Amersham) (D). (A) DBP was stained with rabbit anti-DBP antiserum (20); early gene expression was analyzed with rabbit antisera raised against LEF-3 (3), IE2 (13), LEF-4 (11), or P35 (9); and expression of the cellular TBP was determined with rabbit anti-Sf/TBP antiserum (clone 3890; I. Quadt, unpublished results). (B) Three independent experiments were quantified by ImageJ software, and the increase (fold) in signal intensity in cells with suppressed DBP compared to the level in control cells is given. (C) Structural gene expression was analyzed with anti-GP64 (AcV5) (10) or anti-P39 (P10C6) (25) mouse monoclonal antibodies. (D) Expression of the very late protein P10 was detectable with rabbit anti-P10 serum (23). (E) Polyhedrin was viewed by Ponceau staining. Expression of DBP, LEF-3, IE2, TBP, LEF-4, P39, P10, and polyhedrin was analyzed on samples prepared from nuclear protein extracts, and P35 and GP64 expression was detected in cytosolic extracts that were prepared as described previously (19). Protein size markers are shown on the left, and the identities of the viral proteins are indicated on the right.