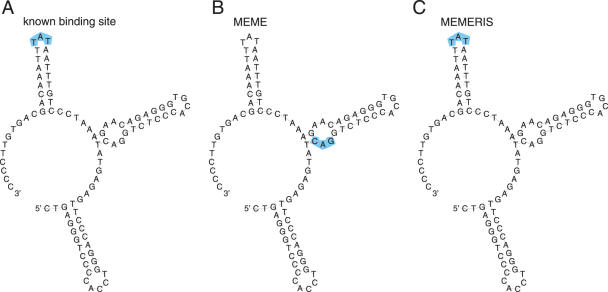
Figure 8.
Results of MEME and MEMERIS for the SLDE Rfam (RF00183) dataset. The figure shows the consensus sequence and structure of the SLDE element. The hairpin loop of the essential third stem is bound by an unknown protein factor (A). MEME detects a CAG motif which does not overlap the binding site (B). In contrast, MEMERIS identifies the TAT sequence of the hairpin loop as the motif (C). Both motif matrices have an information content of 6 bits. The known binding sites and the predicted motifs are highlighted in blue. The motif length was set to 3 nt. For MEMERIS, the PU values were used with a pseudocount of 0.01.