Figure 1.
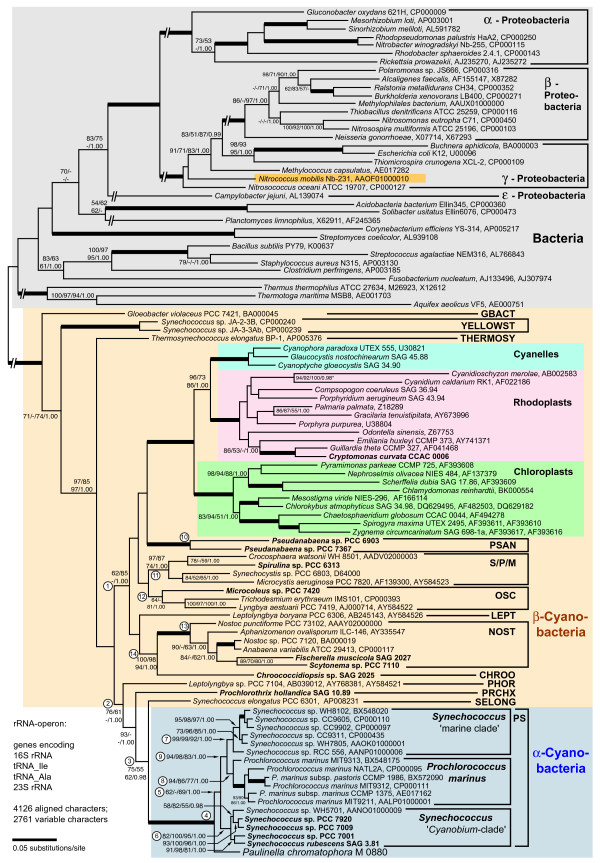
Phylogenetic position of the Paulinella chromatophore within the cyanobacteria inferred by complete rRNA operon sequence comparisons. The tree topology was generated by maximum likelihood (ML) analyses using the GTR+I+Γ model. The nodal support values are bootstrap values ≥ 50% obtained by ML (100 replicates), neighbor-joining (NJ; GTR+I+Γ model; 1000 repl.), maximum parsimony (MP; 1000 repl.), and Bayesian posterior probabilities (≥ 0.95). Branches in bold have maximal support (100%; 1.00) by all methods; interrupted branches were graphically reduced to 30% of their original length. Taxa in bold were newly determined for this study; strain designations and EMBL/GENBANK accession numbers are also given. Cyanobacterial clade abbreviations: GBACT (Gloeobacter), PSAN (Pseudanabaena), S/P/M (Synechocystis/Pleurocapsa/Microcystis), OSC (Oscillatoria), LEPT (Leptolyngbya), NOST (Nostoc), and PHOR (Phormidium): modified sequence groups after [17]; YELLOST (thermophilic "Synechococcus" from Yellowstone NP), THERMOSY (Thermosynechococcus), CHROO (Chroococcidiopsis), PRCHX (Prochlorothrix), SELONG (Synechococcus elongatus), and PS (Prochlorococcus/Synechococcus). Encircled numbers indicate clades/branches that were analyzed by single-gene analyses, and by NJ using the LogDet+I-model (see text and Additional File 1).
