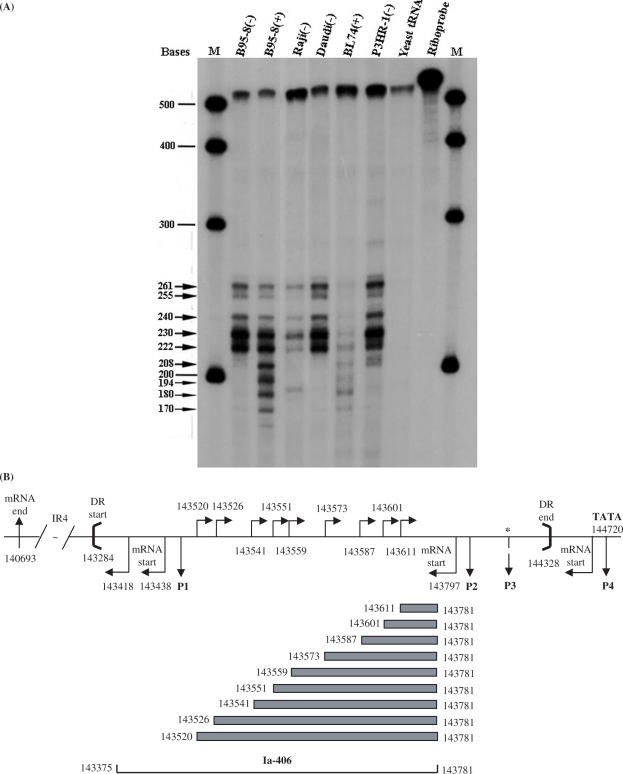
Figure 8.
Identification of complementary (rightward) strand transcripts by RNase protection (RPA) assays. (A) A 32P-radiolabelled 520 base riboprobe (Ia-406, see track, Riboprobe) from the DR region of the viral genome was used in RPA assays to explore ‘rightward’ transcription on the strand complementary to LF3, that is, in a ‘rightward’ direction on the EBV genome (Figure 1). In cells, as noted, a series of small, overlapping products were observed as seen here. Due to deletions in their respective genomes, bands in B95-8 cells must arise from the DL region and in Daudi cells from DR. Raji, BL74 or P3HR-1 bands may represent mixtures of products from both DL and DR. Notably, the patterns are identical, although band intensities vary. For BL74, bands were only observed in induced cells, as shown, which was not the case with other lines. Data given here use a low-specific activity probe, in great excess. Larger bands appear more prominent in uninduced cells, with induction appearing to favour in general the smaller bands. M = molecular weight markers; size calculations of protected bands are given (arrows). (B) An illustrative diagram showing location and nucleotide numbers of the riboprobe (Ia-406) and the protected products from the rightward ‘complementary strand’ transcripts, relative to the BamHI Ia fragment in the EBV genome (33) and DR (between heavy brackets). The locations of the leftward promoter-related sequences, P1–P4 (single-headed arrows) and LF3 mRNA start sites (bent arrows, below horizontal line), both below the horizontal line, are shown. The 5′-end initiation sites of ‘rightward’ transcripts over this same region are indicated (bent arrows) above the horizon line. If expressed simultaneously in cells, the rightward strand transcripts would overlap leftward transcripts, possibly to form double-stranded RNAs which, as such, might be targeted for degradation by cellular enzymes. (*) = mapped only approximately.