Abstract
The Escherichia coli trmA gene encodes the tRNA(m5U54)methyltransferase, which catalyses the formation of m5U54 in tRNA. During the synthesis of m5U54, a covalent 62-kDa TrmA-tRNA intermediate is formed between the amino acid C324 of the enzyme and the 6-carbon of uracil. We have analysed the formation of this TrmA-tRNA intermediate and m5U54 in vivo, using mutants with altered TrmA. We show that the amino acids F188, Q190, G220, D299, R302, C324 and E358, conserved in the C-terminal catalytic domain of several RNA(m5U)methyltransferases of the COG2265 family, are important for the formation of the TrmA-tRNA intermediate and/or the enzymatic activity. These amino acids seem to have the same function as the ones present in the catalytic domain of RumA, whose structure is known, and which catalyses the formation of m5U in position 1939 of E. coli 23 S rRNA. We propose that the unusually high in vivo level of the TrmA-tRNA intermediate in wild-type cells may be due to a suboptimal cellular concentration of SAM, which is required to resolve this intermediate. Our results are consistent with the modular evolution of RNA(m5U)methyltransferases, in which the specificity of the enzymatic reaction is achieved by combining the conserved catalytic domain with different RNA-binding domains.
INTRODUCTION
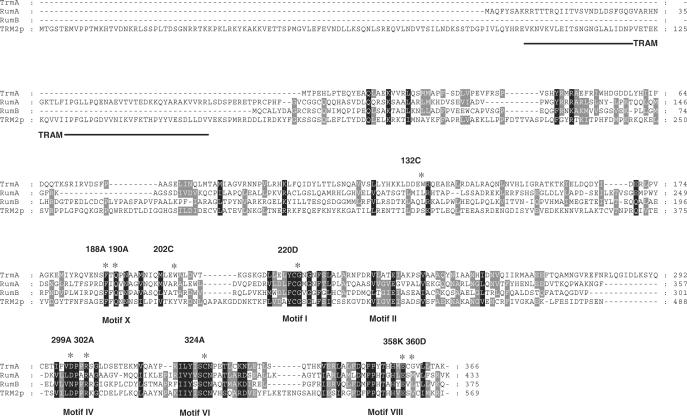
Posttranscriptional RNA modifications appear to be present in all organisms. At present, 107 different types of nucleoside modifications have been established, and 91 of them are found in tRNA (1), (http://medlib.med.utah.edu/RNAmods). One of the most prevalent modified nucleosides found in tRNA is 5-methyluridine (m5U or rT), and in Escherichia coli it occurs once in every tRNA species. The enzyme responsible for this modification in E. coli is encoded by the trmA gene (2). Although m5U is present at position 54 in the TΨC-loop in almost all tRNAs from bacteria and eukarya, its absence induces only a minor growth defect (3,4). The TrmA enzyme belongs to a family of methyltransferases that catalyses methyl group transfer from S-adenosyl-l-methionine (SAM) to position 5 of the heterocyclic base of uridine (U) at position 54 of the tRNA. At present, this family of methyltransferases includes 67 proteins from 42 species, and is listed as COG2265 (clusters of orthologous groups (COGs) (5). The biochemical function of the TrmA, Trm2p, RumA and RumB proteins of COG2265 is known. Both TrmA of E. coli and Trm2p of the budding yeast Saccharomyces cerevisiae catalyse the formation of m5U54 in all tRNA species, except for the yeast initiator tRNAMet (2,4). The RumA and RumB from E. coli synthesize m5U1939 and m5U747 in 23S rRNA, respectively (6,7). Ten different conserved motifs (I-X) are present in the Rossman fold MTases (8), although not all MTases contain all of these motifs. Alignment of the four m5U-forming enzymes TrmA, Trm2p, RumA and RumB reveals six of the ten conserved motifs (Motifs I, II, IV, VI, VIII and X, Figure 1).
Figure 1.
Sequence alignment of four RNA(m5U)methyltransferases with known biochemical function. The conserved motifs, TRAM domain and amino acid substitutions (asterisk, number and nature of the amino acid) investigated in this work are marked. The translational start of the Trm2p is according to (4).
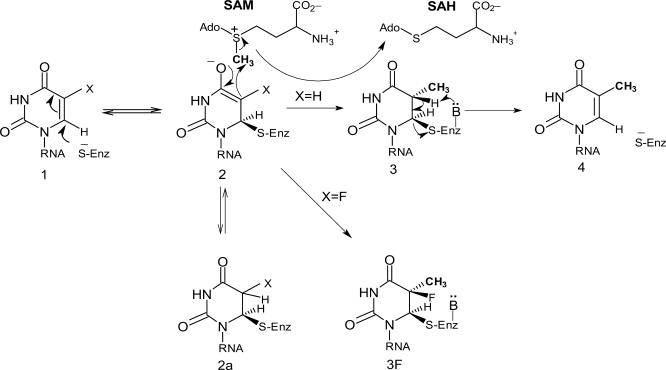
Formation of the m5U54 by TrmA involves a covalent intermediate between the tRNA and a nucleophilic C324 in the enzyme (9). The SH group of C324 reacts with the 6-carbon of U54 in tRNA, producing a nucleophilic centre at the 5-carbon of the U54 (enol or enolate; compound 2 in Figure 2). The methyl group from SAM is transferred to the 5-carbon of U54 (compound 3). Following a β-elimination, m5U54 and the free enzyme (compound 4) are produced. The release of TrmA from the tRNA requires a general base, which has not been identified for TrmA. The U54 is buried in the tRNA through stacking between G53 and Ψ55, and is also involved in a reverse Hoogsteen hydrogen bond with A58. Therefore, prior to catalysis, TrmA must open the T-loop in order to gain access to U54, perhaps by disrupting the hydrogen bonds between the D- and TΨC-arms, which would also disrupt the U54–A58 interaction. This conformational change of the TΨC-loop occurs before the formation of the C324–U54 covalent adduct (9,10). A ‘flip-out’ mechanism similar to that shown for the RumA enzyme is most likely to occur (11).
Figure 2.
The proposed catalytic mechanism of RNA m5U methyltransferases.
The RumA catalyses the formation of the m5U at position 1939 in 23 S rRNA. Its 3D structure has been determined, alone (12) and in complex with RNA and S-adenosyl-l-homocysteine (SAH), the product of the SAM cofactor following transfer of the methyl group to the RNA (11). The catalytic C389 of RumA is present in motif VI as is the catalytic C324 of TrmA (Figure 1). Based on the crystal structure and enzymatic assays of mutant RumA proteins, roles for several additional amino acids in the active site were proposed (11). The F263 (F188 in TrmA) and Q265 (Q190 in TrmA), present in motif X, are important for the U1939 recognition. The D363 (D299 in TrmA) binds to SAH, Q265 and U1939. Amino acid R366 (R302 in TrmA) is also involved in the U1939 binding. The E424 in motif VIII (E358 in TrmA) acts either as the general base releasing the peptide from the enzyme and/or stabilizing the enolate intermediate.
Purification of the TrmA from E. coli revealed that not only the native 42-kDa polypeptide was obtained but also that the native TrmA is associated with RNA (13). The RNA is bound covalently to the enzyme, forming either a 54-kDa complex containing a piece of the 3'-end of 16S rRNA, or a 62-kDa complex containing a subset of undermodified tRNAs (14). The latter complex was suggested to be the TrmA-tRNA intermediate during the formation of m5U54 in tRNA (intermediate 2 in Figure 2). The reason for the presence of the TrmA-16S rRNA linkage is not understood. Thus, in logarithmically growing cells, the enzyme is present in three forms: a 42-kDa native form, a 54-kDa TrmA-rRNA complex and a 62-kDa TrmA-tRNA complex.
Here, we have analysed the formation of m5U54 in tRNA and the formation of the TrmA-tRNA intermediate in several trmA mutants. The mutants were isolated for their inability to make m5U54 in tRNAs (2) and by in vitro mutagenesis. The analysis was made in vivo in exponentially growing cells having the mutated trmA gene in its normal location on the chromosome and with normal levels of the enzyme, SAM and the various tRNA species. Based on the recent findings on the action of the RumA protein (see above), we discuss the role which various amino acids might have in the formation of the TrmA-tRNA intermediate and m5U54 in tRNA. We also compare the role of these amino acids with the corresponding amino acids of RumA. Our results suggest that the conserved amino acids in the TrmA protein, most likely, have similar roles as in RumA. Therefore, the structure of TrmA in the regions important for catalysis is predicted to be similar to that present in RumA. The surprisingly high level of the 62-kDa TrmA-tRNA intermediate found in exponentially growing cells is also discussed, and is suggested to be caused by the suboptimal concentration of SAM, which is required for the resolution of this intermediate.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
Escherichia coli strains and plasmids used are listed in Table 1. LB medium (15) was used for growth of bacteria. When required, carbenicillin and chloramphenicol were used at concentrations of 50 and 15 μg/ml, respectively.
Table 1.
E. coli strains and plasmids
| Strains | Relevant genotype or phenotype | Source/reference |
|---|---|---|
| GB1-5-41 | trmA+ arg | (3) |
| GB1-4-IB | trmA4 (W202C) arg ampA1 | (3) |
| GB1-5-39 | trmA5 (G220D) arg | (3) |
| GB1-6-1 | trmA6 (W132C) met | (3) |
| GB1-9-6 | trmA9 (G360D) | (3) |
| GB1-10-4 | trmA10 (E358K) met thiA | (3) |
| MW100 | trmA+ | M. Wikström |
| GRB2268 | MW100/pKD46 | M. Wikström |
| GRB1648 | trmA17(C324A) | This work |
| GRB2269 | yijD::kan | This work |
| GRB2279 | yijD::kan trmA14 (D299A) | This work |
| GRB2293 | yijD::kan trmA15 (F188A) | This work |
| GRB2294 | yijD::kan trmA16 (Q190A) | This work |
| GRB2230 | yijD::kan trmA18 (R302A) | This work |
| SY327 λpir | Δ(lac-pro) argE(Am) rif nalA recA56 λpir | (39) |
| Plasmids | ||
| pGP100 | trmA+ CbR | (17) |
| pGP100 C324ATrmA | trmA17(C324A) CbR | This work |
| pJU3 | sacB trmA17 (C324A) CmR | This work |
| pDM4 | SacB CmR | (18) |
| pKD4 | KmR | (20) |
| pKD46 | CbR | (20) |
DNA manipulations
Procedures for DNA digestions, agarose gel electrophoresis, DNA ligation and transformation of competent E. coli cells were performed essentially as described earlier (16).
Amplification of DNA by PCR
The PCR amplification was performed using Taq DNA polymerase (Boehringer Mannheim GmbH, Mannheim, Germany) using the buffers supplied with the enzymes. Routinely, 5 pmol of the appropriate primers and ∼100 ng template DNA were added to the reaction mixture. Alternatively, the trmA gene was amplified from cell suspensions using the PuReTaq Ready-To-Go™ PCR Beads (Amersham, UK) and purified by the PCR Kleen Spin Kit (Biorad). The PCR products were visualized by running 1% agarose gels, staining with ethidium bromide and exposition to the UV light.
Construction of the mutants
In vitro mutagenesis to obtain the C324A substitution in the TrmA protein was done on the pGP100 plasmid (17) using QuickChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA, US) according to the instructions of the manufacturer. The mutated trmA gene was moved to the pDM4 suicide plasmid (18) which subsequently was transformed into the strain MW100. Resulting duplications were resolved by growing cells in the presence of 5% sucrose. The duplications are resolved since the sucrose is toxic for E. coli containing the pDM4 with sacB gene (19). Presence of the mutation corresponding to the mutant C324A TrmA was verified by sequencing.
Alternatively, in order to mutate the other codons in the trmA gene, a kanamycin resistance cassette from the plasmid pKD4 was placed between codons 107 and 108 of the yijD gene by linear transformation into strain MW100 (20). Since yijD gene is close to the trmA gene on the chromosome, this strain was used as a template in a PCR, where one of the primers was homologous to the kanamycin resistance cassette, and the other contained a desired mutation in the trmA gene. The resulting product containing the resistance gene and the mutation was then transformed into strain MW100 carrying the pKD46 plasmid coding for the λ Red recombinase. The transformants were screened for the desired mutations by sequencing.
DNA sequencing and sequence analysis
Column-purified PCR fragments were used to sequence mutations in the trmA gene. Sequencing was mainly performed with a BigDye Ready Reaction Kit (Perkin Elmer) sequencing premix in an ABI Prism 377 DNA sequencer. The sequences were analysed using the nucleotide BLAST at the National Center for Biotechnology Information (www.ncbi.nml.nih.gov/blast).
Analysis of tRNA modification levels by HPLC
Different trmA mutants were grown in LB medium at 37°C to ∼4 × 108 cells/ml and harvested. Transfer RNA was prepared as described previously (21) and degraded to nucleosides with P1 nuclease followed by treatment with alkaline phosphatase (22). The hydrolysate was analysed by high-performance liquid chromatography (HPLC) (23) on a Supelcosil LC18 column (Supelco) with Waters HPLC system. Alternatively, the hydrolysates were run on a Develosil 5µ RP-AQUEOUS C30 column (Phenomenex) with an identical gradient. The level of m5U modification was normalized to the absorbance of t6A at 254 nm. The relative amounts of m5U in each mutant varied ± 15% in different runs. The detection limit was calculated by comparing the area of a small clearly visible peak to the area of t6A.
Immunoblotting
Bacteria were grown in 10 ml LB broth to a density of ∼4 × 108 cells/ml. The cells were disrupted by sonication, and cell debris was removed by centrifugation. Twenty micrograms of protein from the supernatant was separated by 12% SDS-PAGE. Separated proteins were blotted onto a Hybond-C™ membrane (Amersham Life Science, UK) essentially as described by (24) and immunodetection was performed using the ECL-PLUS western blotting kit (Amersham Life Science, UK). Primary antibodies, specific for the m5U54-methyltransferase, were a kind gift from D. Santi (San Francisco, CA, US). Bands were scanned using a Fluor-S™ MultiImager (Biorad, Hercules, CA, US) and quantified using the Quantity One® software. The relative intensities of the TrmA proteins varied ± 15% in different western blots. We suggest that several additional bands appearing on the western blots is cross-reacting material since they are present in an E. coli strain deleted for the trmA gene and in the trmA4 mutant containing no detectable TrmA protein (see Results).
Sequence analysis
BLAST program (25) was used to search for the gene and protein sequences, mainly at NCBI. The TrmA protein family was analysed using the cluster of orthologous groups at www.ncbi.nih.gov/COG/. Sequences were aligned using Multalin program (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html) (26), and the alignments were manipulated manually using the Genedoc program (http://www.psc.edu/biomed/genedoc).
RESULTS
Alignment of the TrmA family of m5U-methyltransferases with a known function
Four proteins of the TrmA family with known biochemical function were aligned using the Multalin program. They display several well-established motifs typical to the Rossman-fold-like SAM-dependent methyltransferases (Figure 1) (8). The S. cerevisiae tRNA(m5U54)methyltransferase (Trm2p) has a long N-terminal extension, which is absent in the E. coli enzymes. The 23 S rRNA(m5U)methyltransferases RumA and RumB are characterized by the presence of an [Fe4S4] cluster-binding motif (C81, C87, C90 and C162, RumA nomenclature). The presence of such a cluster was experimentally demonstrated for RumA (27), but it is not clear whether it is present in RumB. No [Fe4S4] cluster-binding motif is present in TrmA and Trm2p. Further, TRAM, a predicted RNA-binding domain, is present in the N-terminus of RumA and of Trm2p, but is lacking in the TrmA and RumB proteins.
Steady-state levels of m5U54 in tRNA and of the TrmA-tRNA intermediate in trmA mutants randomly isolated as being deficient in m5U54 in tRNA
Several trmA mutants were randomly isolated as being deficient in m5U54 in their tRNA (2). The amino acid changes in these mutants have been established by sequencing and are marked by ‘*’ in Figure 1 and the corresponding strains listed in Table 1. Relative amounts of the native 42-kDa TrmA enzyme and the 62-kDa TrmA-tRNA intermediate were determined in the various mutants by western blot analysis. From the same cultures, the level of m5U54 in tRNA was also measured by HPLC analysis. All cultures were grown in LB medium at 37°C and harvested at a cell density of 4 × 108 cells/ml to ensure that the cells were in the exponential growth phase. The results are presented in Figure 3.
Figure 3.
Western blot analysis of trmA mutants. Strains MW100 (wt), GB1-41B (trmA4), GB1-5-39 (trmA5), GB1-6-1 (trmA6), GB1-9-6 (trmA9), GB1-10-4 (trmA10), GRB2293 (trmA15), GRB2294 (trmA16) and GRB1648 (trmA17) were used to prepare the protein extracts. The intensities of the 42- and 62-kDa peptides of wild type and of each mutant are expressed as a percentage of the total intensities of these peptides (42 + 62 kDa; total intensity of 100%, which is not shown). The row labelled ‘TrmA,%wt’ shows the level of total TrmA-associated peptides in the various mutants relative to the level found in wild type. The ‘x’ band was used as an internal control for the amount of the loaded protein extracts. The row labelled ‘m5U,%wt (HPLC)’ shows the amount of m5U in total tRNA in each mutant expressed as a percentage of the level of m5U in tRNA from the wild-type strain. The asterisk indicates that the amount of m5U from the HPLC chromatogram is an overestimate due to impurities in the peak.
The trmA4 mutant has a W202C amino acid exchange in TrmA, but also a silent mutation in a codon corresponding to H340 at the C-terminus. This mutant has only 15% of the wild-type level of the m5U54 in tRNA, and no detectable native TrmA or TrmA-tRNA intermediate. Apparently, the residual level of enzyme present in the cell is sufficient to catalyse the formation of some m5U54 in tRNA, but the enzyme is quickly degraded before or during protein extraction for western blot analysis. The G220D amino acid substitution in the putative SAM-binding site in the trmA5 mutant results in almost no m5U54 in tRNA and no 62-kDa TrmA-tRNA intermediate. The trmA6 mutant has a W132C substitution, which leads to an increased level of the 62-kDa TrmA-tRNA intermediate and a 66% decreased level of m5U54, suggesting that this alteration in TrmA decreases the resolution of the intermediate, resulting in less formation of m5U54. Two other mutants, trmA9 and trmA10, contain G360D and G358K substitutions, respectively, located at the extreme C-terminus of the TrmA. These mutants do not accumulate the 62-kDa TrmA-tRNA intermediate, and have a very much reduced level of m5U54. Since the total level of TrmA was reduced to about 55% of that found in the wild type, these alterations probably also reduce the stability of the TrmA protein.
Levels of m5U54 in tRNA and the TrmA-tRNA intermediate in trmA mutants created by site-directed in vitro mutagenesis
In order to test which additional amino acids could be important for the formation of the covalent tRNA-TrmA complex and the formation of m5U54 in tRNA, several additional alleles of the trmA gene in strain MW100 were created (Table 1). Motif X of the TrmA protein contains two conserved residues, F188 and Q190, predicted to have an important role for the uracil recognition of the RNA(m5U)methyltransferases (corresponding to the F263 and Q265 in RumA, see above). We have constructed mutants trmA15 and trmA16, which have the F188A and Q190A alterations in TrmA, respectively. Although the combined level of the native TrmA and the TrmA-tRNA intermediate was only slightly reduced to 84% of the wild-type level in the F188A mutant, the ratio between these two forms was about the same as in wild-type cells. However, the level of m5U54 in tRNA was only 22% of that found in wild-type strain. By contrast, a Q190A substitution, which is located close to the F188, leads to almost no formation of the TrmA-tRNA intermediate, although the total level of the TrmA proteins were about the same as in the trmA15 (F188A) mutant (77%). The m5U54 level was reduced to 14% of wild-type level in the Q190A mutant. Apparently, the Q190A alteration, but not the F188A alteration, affects the step resulting in the formation of the TrmA-tRNA intermediate, which in turn is pivotal for the formation of m5U54 in tRNA according to the model of catalysis (Figure 2).
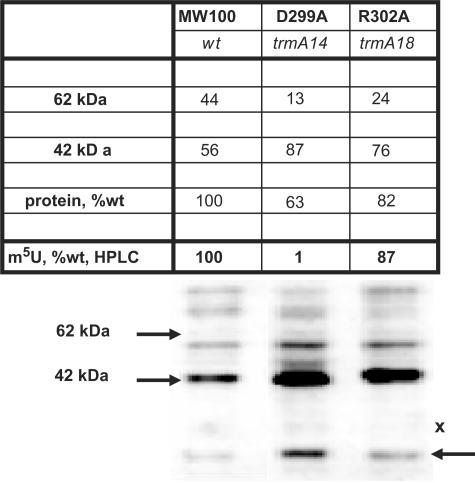
The D363 and R366 residues in the motif IV of RumA were proposed to bind the U1939 of 23 S rRNA (11,12,28). To test the significance of the corresponding residues in TrmA (D299 and R302, respectively), alterations in these residues were obtained by site-directed mutagenesis, resulting in the chromosomal trmA14 (D299A) and trmA18 (R302A) mutants (corresponding to D363 and R366 in RumA). In the trmA14 (D299A) mutant, the level of the 62-kDa tRNA-TrmA intermediate is significantly reduced, and accordingly, the level of m5U54 in tRNA is almost undetectable (Figure 4). In the trmA18 construct, the level of the 62-kDa TrmA-tRNA intermediate is significantly reduced, but the amount of the m5U54 in tRNA is only slightly reduced to 87% of the wild-type level. Apparently, these two amino acids, being close to each other, have a very different impact on the activity of the TrmA in vivo.
Figure 4.
Western blot analysis of trmA mutants in motif IV. Strains MW100 (wt), GRB2276 (trmA14) and GRB2230 (trmA18) were used to prepare the protein extracts. The values were calculated as in Figure 3.
Amino acid C324 was demonstrated to be the catalytic amino acid residue in vitro (9). We have therefore created the chromosomal trmA17 allele encoding the C324A substitution in TrmA by directed in vitro mutagenesis. As expected, this mutant does not form the 62-kDa tRNA-TrmA complex, and contains no detectable m5U54 in the tRNA (Figure 3). Note that the level of the native TrmA was also reduced, indicating that this enzyme is less stable when it is not able to bind to tRNA.
DISCUSSION
We show here how various amino acid alterations of the tRNA(m5U54)methyltransferase (TrmA) affect the synthezsis of m5U54 in tRNA, and how the relative levels of the 42-kDa native form of TrmA and the 62-kDa TrmA-tRNA intermediate were affected in vivo. Amino acid substitutions of F188, Q190, G220, D299, R302, C324 and E358, which are conserved in the four biochemically characterized RNA(m5U)methyltransferases (Figure 1), reduce the formation of the covalent 62-kDa tRNA-TrmA intermediate and/or the enzymatic activity, as shown by the reduced level of m5U54 in tRNA. Also, the substitutions of W132 or W202, conserved among the bacterial tRNA(m5U54)-methyltransferases but not among the other RNA(m5U)methyltransferases, reduce the synthesis of m5U54 in tRNA. Moreover, the W202 is important for the stability of TrmA, and the W132C alteration resulted in increased accumulation of the 62-kDa TrmA-tRNA intermediate as compared to the wild-type level of this intermediate. Our results suggest that the structural elements important for activity of TrmA are similar to those of RumA, which is responsible for the formation of the same methylated nucleoside in position 1939 of 23S rRNA, and for which the 3D structure of the catalytic centre is known.
Here, we have analysed the formation of the TrmA-tRNA intermediate and the m5U54 in tRNA in several trmA mutants. Our analysis was performed in true in vivo conditions, since the TrmA protein was expressed from the trmA gene located at its normal position on the chromosome. Thus, unlike the in vitro approach, our method compares the enzymatic activity of the wild-type and mutant tRNA(m5U54)methyltransferases at physiologically normal conditions. In addition, the relative level of the mutant forms of the TrmA enzyme and formation of the 62-kDa TrmA-tRNA reaction intermediate were monitored using the same batch of cells. We believe, therefore, that the results obtained by such analysis reflect the kinetics of m5U54 formation in tRNA in normal in vivo conditions.
We discuss below the influence of various amino acids in the TrmA protein on the formation of m5U54 in tRNA in relation to those important for the synthesis of m5U1939 in 23S rRNA catalysed by RumA protein as suggested by an analysis of the 3D structure of this enzyme. The crystal structure of the RumA protein was determined alone (12) and in complex with RNA and the inhibitor SAH (11). Although the functions of several conserved amino acids in RumA were proposed (see Introduction), the enzymatic activities of mutant RumA proteins were experimentally tested only for alterations of Q265 (corresponding to Q190 in TrmA), D363 (D299 in TrmA) and E424 (E358 in TrmA) (11). Here, we have tested the enzymatic activity of the corresponding amino acids in TrmA, as well as several other amino acids judged to be important for catalysis.
Amino acid C324 of the TrmA is the catalytic amino acid residue in vitro (9). The corresponding C389 of the RumA is covalently attached to carbon-6 of U1939 via a thioether linkage in the RumA-23S rRNA co-crystal (11), Figure 5A). The C324A TrmA mutant is unable to form the covalent TrmA-tRNA intermediate and m5U54 in tRNA (Figure 3), which confirms the crucial role played in catalysis by the corresponding cysteines in RumA and TrmA, respectively. Moreover, the absence of the 62-kDa TrmA-tRNA complex in this mutant is consistent with our suggestion that this complex is the postulated intermediate compound 2 or 2a (Figure 2).
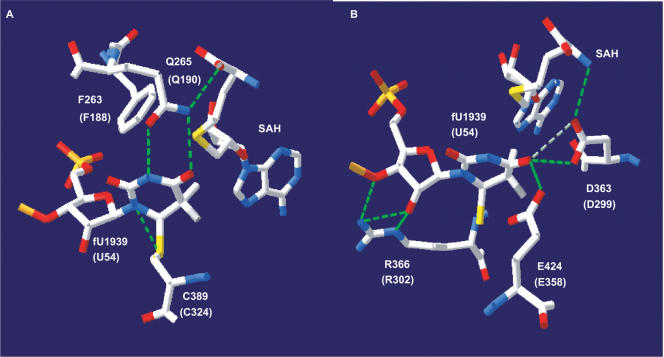
Figure 5.
Interactions between amino acid chains, the target uridine and SAH in the active site of RumA. Several selected amino acids in the active site are divided into (A) and (B) for better representation. Corresponding residues in the TrmA are shown within the parenthesis. The image was created with the SwissPDB Viewer program (40) http://www.expasy.org/spdbv/) using the coordinates of RumA-RNA-SAH complex (2BH2) (11) from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). Putative hydrogen bonds are represented by dashed lines.
The F188A amino acid substitution in motif X of TrmA did not affect the level of the 42-kDa native TrmA or of the 62-kDa TrmA-tRNA intermediate but severely reduced the synthesis of m5U54 in tRNA (Figure 3). The corresponding F263 of RumA forms an edge-to-face aromatic interaction with the uracil ring and is itself held by the sugar-phosphate backbone of U1939 and the homocysteine moiety of SAH [Figure 5, (11)]. Since the F188A mutation does not affect the formation of the 62-kDa TrmA-tRNA intermediate (Figure 3), it suggests that F188 (F263 in RumA) is not important for the positioning of the uracil ring and the formation of the intermediates 2 and 2a (Figure 2). Instead, it may be important for the proper positioning of SAM for the methylation of the U54. In contrast, the Q190A substitution in TrmA affects both the formation of the covalent 62-kDa TrmA-tRNA complex and the synthesis of m5U54 in tRNA (Figure 3). Q265 (Q190 in TrmA) in RumA forms hydrogen bonds with N3 and with O4 of U1939 (Figure 5), and is also involved in the binding of SAM. The Q265A mutant of RumA displays an 830-fold lower specific activity compared to the wild-type enzyme (11). These and our results suggest that Q190 (Q265 of RumA) is the primary uracil-recognizing residue, and is important for positioning of the U target prior to the nucleophilic attack by the catalytic cysteine.
The D299A alteration in motif IV of TrmA leads to a severely reduced formation of the TrmA-tRNA intermediate and absence of m5U54 in tRNA (Figure 4). The corresponding D363A substitution in RumA also results in complete loss of the RumA activity (11). According to the 3D structure of RumA, D363 makes two H-bonds with O4 of U1939 and one H-bond with SAH (Figure 5B). We propose that D299 has a similar role in TrmA. The guanidine group of R366 in the motif IV of RumA hydrogen bonds to O2′ and O3′ of the ribose of U1939 (Figure 5B), which suggests an important role for this amino acid residue in RumA. However, the R302A (R366 in RumA) alteration in TrmA only slightly reduced the formation of m5U54, although the formation of the TrmA-tRNA intermediate was reduced by about 2-fold (Figure 4). Apparently, the role of R302 in the formation of m5U54 is only a minor one, and the lower level of the TrmA-tRNA intermediate in the R302A mutant is likely due to reduced stability during cell extraction for western blot analysis.
The E424 of RumA was demonstrated to be a general base for proton abstraction and ß-elimination, since intermediate 3 (Figure 2) accumulated in the presence of SAM in a reaction catalysed by an E424Q mutant (11). One would expect that in the absence of SAM, such a mutant enzyme should catalyse the formation of intermediate 2 and/or 2a. However, this is not the case because the E424Q alteration probably affects the relative stabilities of the reaction intermediates 2 and 2a (Figure 2), possibly by changing the local electrostatic environment. The corresponding E358K mutant of TrmA has a very low level of the TrmA-tRNA reaction intermediate, and the cellular level of m5U54 is only 9% of the wild-type strain. We therefore suggest that E358 is the general base for proton abstraction and ß-elimination in TrmA. As in the case of RumA, substitution of E358 probably makes the reaction intermediates 2 and 2a unstable.
The trmA5 (G220D) mutation that alters the putative SAM-binding site and thereby abolishing the formation of m5U54, also abolished the formation of the 62-kDa TrmA-tRNA complex (Figure 3), which we suggest to be the reaction intermediate 2 and/or 2a (see above). Accumulation of this reaction intermediate(s) occurs in the absence of SAM (29), while one would expect its accumulation in the mutant deficient in SAM binding. However, this is not the case. Apparently the G220D alteration, which probably changes the structure of the SAM-binding domain, also blocks the formation of the covalent intermediate 2 and/or 2a, similarly to the E358K mutant (and E424Q mutant of RumA).
About 30–45% of the wild-type TrmA protein in E. coli is covalently bound to undermodified tRNA as the 62-kDa TrmA-tRNA complex [Figures 3 and 4; (14)], which represents the intermediates 2 and 2a (Figure 2). The resolution of intermediate 2 requires SAM (29). Therefore, presence of this intermediate at such high level in exponentially growing wild-type cells may indicate that the concentration of SAM is suboptimal in relation to the Km of TrmA, which is between 12.5 (30) and 17 μM (13). When E. coli is growing exponentially in LB at 37°C (i.e. the same conditions as used by us), the intracellular concentration of SAM is between 1 μM (31) and 13 μM (32) assuming a cell volume of ∼2 × 10−15 l (33). Thus, the estimated intracellular level of SAM is below the required concentration for attaining Vmax for TrmA to resolve the 62-kDa TrmA-tRNA intermediate. This may explain the relatively high level of such intermediate in wild-type cells. Interestingly, the cellular level of the dimethylallyl pyrophosphate, the cofactor for the production of another tRNA modification, i6A37, is limited. This results in a reduced level of i6A37 if the demand for dimethylallyl pyrophosphate is increased in other areas of metabolism in which it also participates (34). Since some hypomodification results in less efficient translation, these cases exemplify links between metabolism and translation, and may constitute a regulatory device for their co-ordination (35,36).
Some of the trmA mutants were isolated by the classical genetic approach of random screening for the mutants unable to form m5U54 in tRNA (2). Although such an approach resulted in alterations in well-established sequence motifs (trmA5 and trmA10) and their function could be predicted by the ‘sequence–structure–function’ approach, some of the isolated trmA mutants (trmA4 and trmA6) have alterations in unexpected positions, and their role in the formation of m5U54 cannot be easily explained at present. However, when a 3D structure of TrmA is available, their role in the synthesis of m5U54 should be apparent and validate the structure. These results demonstrate the usefulness of an unbiased genetic approach to elucidate the role of certain amino acids in the protein in addition to the ‘sequence–structure–function’ approach which requires detailed knowledge of the protein structure.
In summary, our results suggest that several conserved amino acids in the C-terminal domain are important for catalysis in both TrmA and RumA proteins. In addition, the G428D or the C521A substitutions (corresponding to the G220D and C324A of TrmA, respectively) completely inactivated Trm2p (37). The TrmA protein lacks the N-terminal ‘TRAM’ domain that is present in Trm2p and is involved in the 23S rRNA binding in RumA [Figure 1; (11)]. The RNA substrate recognition should therefore be different between TrmA and the two other RNA(m5U)methyltransferases, even though the catalytic domains seem to share extensive similarity. Our results are consistent with the theory of the modular evolution of RNA-modifying enzymes (38), which suggests that the specificity of the enzymatic reaction is achieved by combining different (predicted) RNA-binding domains with different catalytic domains.
ACKNOWLEDGEMENTS
We thank Drs D. Milton for providing plasmid pDM4, J. Näsvall for the stimulating discussions and help in the preparation of Figure 5. We also thank A. Byström, J. Durand, T. Hagervall, J. Näsvall, O. Persson and I. Tittawella for the critical reading of the manuscript. We also thank I. Tittawella for linguistic improvement of the manuscript. This work was supported by grants to G.R.B. from the Swedish Cancer Foundation (project 680) and Science Research Council (project B-BU 2930). Funding to pay the Open Access publication charges for this article was provided by Swedish Cancer Foundation and Science Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björk GR, Isaksson LA. Isolation of mutants of Escherichia coli lacking 5-methyluracil in transfer ribonucleic acid or 1-methylguanine in ribosomal RNA. J. Mol. Biol. 1970;51:83–100. doi: 10.1016/0022-2836(70)90272-x. [DOI] [PubMed] [Google Scholar]
- 3.Björk GR, Neidhardt FC. Physiological and biochemical studies on the function of 5-methyluridine in the transfer ribonucleic acid of Escherichia coli. J. Bacteriol. 1975;124:99–111. doi: 10.1128/jb.124.1.99-111.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordlund ME, Johansson JOM, von Pawel Rammingen U, Byström AS. Identification of the TRM2 gene encoding the tRNA(m5U54) methyltransferase of Saccharomyces cerevisiae. RNA. 2000;6:844–860. doi: 10.1017/s1355838200992422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 7.Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31:4738–4746. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of AdoMet-dependent methyltransferases. In: Cheng X, Blumenthal RM, editors. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Singapore: World Scientific; 1998. pp. 1–38. [Google Scholar]
- 9.Kealey JT, Santi DV. Identification of the catalytic nucleophile of tRNA (m5U54)methyltransferase. Biochemistry. 1991;30:9724–9728. doi: 10.1021/bi00104a022. [DOI] [PubMed] [Google Scholar]
- 10.Yao LJ, James TL, Kealey JT, Santi DV, Schmitz U. The dynamic NMR structure of the T psi C-loop: implications for the specificity of tRNA methylation. J. Biomol. NMR. 1997;9:229–244. doi: 10.1023/a:1018618606857. [DOI] [PubMed] [Google Scholar]
- 11.Lee TT, Agarwalla S, Stroud RM. A unique RNA fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 12.Lee TT, Agarwalla S, Stroud RM. Crystal structure of RumA, an iron-sulfur cluster containing E. coli ribosomal RNA 5-methyluridine methyltransferase. Structure (Camb.) 2004;12:397–407. doi: 10.1016/j.str.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Ny T, Lindström HR, Hagervall TG, Björk GR. Purification of transfer RNA (m5U54)-methyltransferase from Escherichia coli. Association with RNA. Eur. J. Biochem. 1988;177:467–475. doi: 10.1111/j.1432-1033.1988.tb14396.x. [DOI] [PubMed] [Google Scholar]
- 14.Gustafsson C, Björk GR. The tRNA-(m5U54)-methyltransferase of Escherichia coli is present in two forms in vivo, one of which is present as bound to tRNA and to a 3′-end fragment of 16 S rRNA. J. Biol. Chem. 1993;268:1326–1331. [PubMed] [Google Scholar]
- 15.Bertani G. Studies on lysogenesis. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Lindström PH, Stüber D, Björk GR. Genetic organization and transcription from the gene (trmA) responsible for synthesis of tRNA (uracil-5)-methyltransferase by Escherichia coli. J. Bacteriol. 1985;164:1117–1123. doi: 10.1128/jb.164.3.1117-1123.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Björk GR, Jacobsson K, Nilsson K, Johansson MJO, Byström AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrke CW, Kuo KC, McCune RA, Gerhardt KO, Agris PF. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr. 1982;230:297–308. [PubMed] [Google Scholar]
- 23.Gehrke CW, Kuo KC. Ribonucleoside analysis by reversed-phase high performance liquid chromatography. In: Gehrke CW, Kuo KCT, editors. Chromatography and Modification of Nucleosides. Part A. Analytical Methods for Major and Modified Nucleosides. 1990. Vol. 45A J. Chromatography Library. Elsevier, Amsterdam, pp. A3–A71. [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwalla S, Stroud RM, Gaffney BJ. Redox reactions of the iron-sulfur cluster in a ribosomal RNA methyltransferase, RumA: optical and EPR studies. J. Biol. Chem. 2004;279:34123–34129. doi: 10.1074/jbc.M405702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X, Santi DV. Covalent adducts between tRNA (m5U54)-methyltransferase and RNA substrates. Biochemistry. 1992;31:10295–10302. doi: 10.1021/bi00157a017. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg R, Dudock B. Isolation and characterization of m5U-methyltransferase from Escherichia coli. J. Biol. Chem. 1980;255:8296–8302. [PubMed] [Google Scholar]
- 31.Spoerel N, Herrlich P, Bickle TA. A novel bacteriophage defence mechanism: the anti-restriction protein. Nature. 1979;278:30–34. doi: 10.1038/278030a0. [DOI] [PubMed] [Google Scholar]
- 32.Posnick LM, Samson LD. Influence of S-adenosylmethionine pool size on spontaneous mutation, dam methylation, and cell growth of Escherichia coli. J. Bacteriol. 1999;181:6756–6762. doi: 10.1128/jb.181.21.6756-6762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donachie WD, Robinson AC. Cell division: parameter values and the process. In: Neidhardt FC, Ingraham JL, Brooks LK, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1987. pp. 1578–1593. [Google Scholar]
- 34.Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc. Natl Acad. Sci. USA. 2000;97:61–66. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Björk GR, Rasmuson T. Links between tRNA modification and metabolism and modified nucleosides as tumor marker. In: Grosjean H, Benne B, editors. Modification and Editing of RNA. Washington, DC: American Society for Microbiology; 1998. pp. 471–491. [Google Scholar]
- 36.Björk GR, Nilsson K. 1-Methylguanosine-deficient tRNA of Salmonella enterica Serovar Typhimurium affects thiamine metabolism. J. Bacteriol. 2003;185:750–759. doi: 10.1128/JB.185.3.750-759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson MJ, Byström AS. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA. 2002;8:324–335. doi: 10.1017/s1355838202027851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anantharaman V, Koonin EV, Aravind L. TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol. Lett. 2001;197:215–221. doi: 10.1111/j.1574-6968.2001.tb10606.x. [DOI] [PubMed] [Google Scholar]
- 39.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]