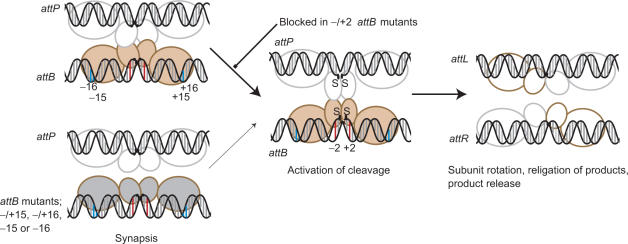
Figure 7.
Model for the mechanism of integrase. The integrase subunits are shown with a small N-terminal (catalytic) domain through which the subunits may dimerize (26) and a large C-terminal domain that we propose recognizes the sequences in the outer flanks of the recombination sites. These recognition events, specifically −15 and −16 (annotated as blue bars) in attB, lead to an ‘induced fit’ or stabilization of a specific conformation of integrase that enables synapsis with integrase bound to attP. Different conformations of integrase bound to either attB or attP are shown as different colours. The synaptic interface via the N-terminal catalytic domains is indicated based on the resolvase precedent; there is no evidence to indicate that the C-terminal domains could also participate in a synaptic interface. Mutations at positions −15 or −16 (such as in T-15C:C+15G, G-16T:G+16A), T-15C or G-16T do not induce the conformation of integrase that can form a stable synapse with attP so the rate of reaction decreases (thin arrow). Mutations at −/+2 in attB (red bars) are severely inhibited in cleavage but are capable of forming a stable synapse. These mutants indicate that the formation of the synaptic complex is followed by a well-defined activation step that results in concerted DNA cleavage. After strand exchange integrase is bound to the hybrid sites and adopts a conformation that cannot synapse attL and attR. The putative tetrameric complex rapidly dissociates to binary complexes containing integrase and either attL or attR. See text for more details.