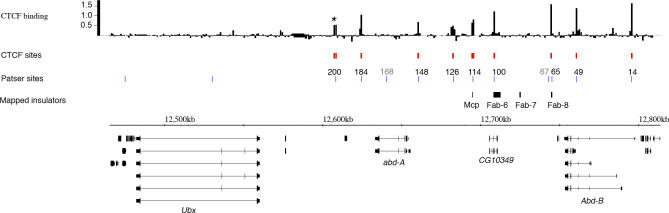
Figure 2. CTCF Binding Profile across the BX-C.
The top track shows the CTCF Mn per fragment across the region. Black asterisk marks fragment UBX200. CTCF sites are sites with Patser p < 10−13, which also show significant enrichment (mean > 0.45; p < 0.05, red bars). Patser sites are positions of Patser matches to CTCF consensus with p < 10−13 (blue bars). The numbers above the blue bars relate the Patser sites to the fragments used in the validation ChIPs and EMSA (Figure 3); the sites 67 and 168 (grey) are not associated with significant enrichment. The positions 148, 100, and 65 have closely spaced double sites. The positions of the mapped insulator elements are indicated above the sequence coordinate line and RefSeq genes below. Enriched fragments correlate well with Patser sites and with the positions of three of the mapped insulators; Mcp, Fab-6, and Fab-8.