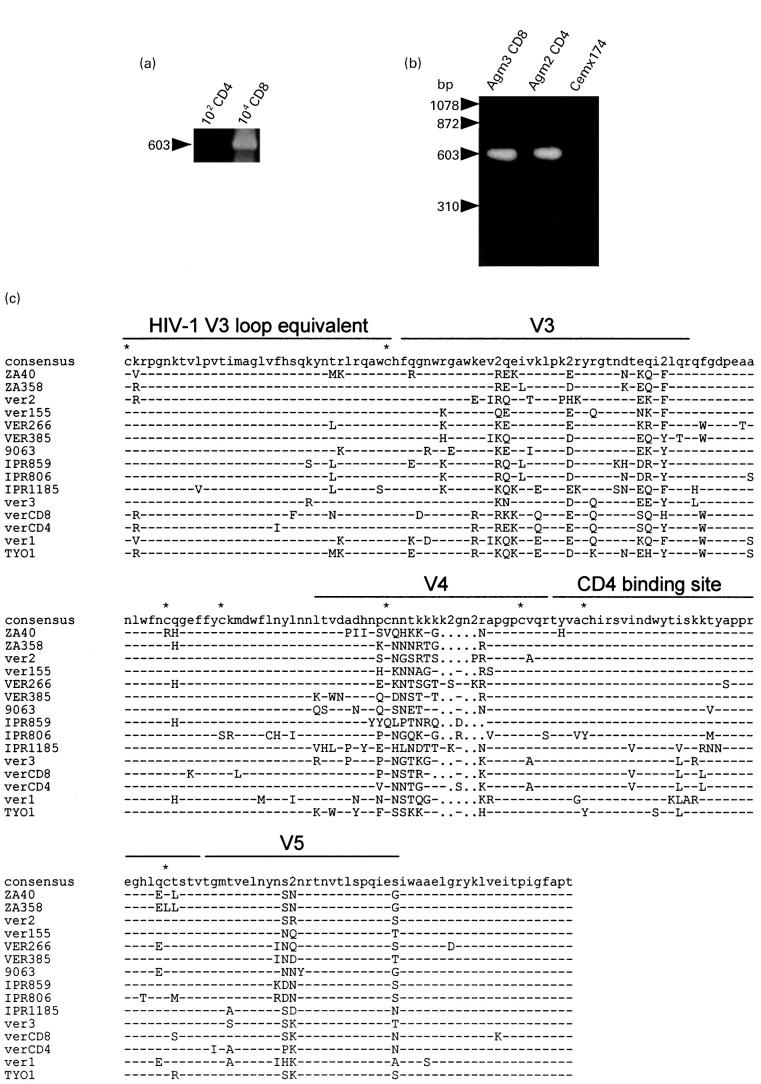
Fig. 2.
(a) Detection of proviral DNA in CD4 and CD8 subsets in Agm3. DNA was extracted from sorted 102–104 cells and employed for nested-polymerase chain reaction (PCR). (b) Detection of SIVagm provirus by the PCR and Southern blotting in Cemx174 cells cocultured with peripheral CD4 or CD8 cells from African green monkeys. A negative reaction was confirmed in the DNA sample of CemX174 cells cultured alone. (c) Multiple alignment of deduced amino acid sequences of SIV isolated from vervet monkeys. PCR derived env nucleotide sequences from Agm2 CD4 cells (verCD4) and Agm3 CD8 cells (verCD8) were translated and aligned with previously reported SIV env sequences. Dashes indicate sequence identity with the consensus sequence, while dots represent gaps introduced to optimize the alignment. ‘2’ in the consensus sequence denotes sites in which two amino acids were shared in viruses at the same frequency. Asterisks indicate cysteine residues. V3, V4 and V5 designate variable regions. The putative CD4 binding site and HIV-1 V3 loop equivalent were as described previously [16,26]. The sequences of the references were obtained from the DNA data bank of Japan (DDBJ): ZA40 (AF015906), ZA358 (AF015905), IPR806 (AF015907), IPR859 (AF015908) and IPR1185 (AF015809) [30]; ver1 (U04003) and ver2 (U04004) [31]; ver155 (M29975) [32]; VER266 (U10896) and VER385 (U10898) [33]; 9063 (L40990) [34]; ver3 (M30931) [35]; and TYO1 (X07805) [1]. 9063 was isolated from a pig-tailed macaque inoculated with SIVagm90.