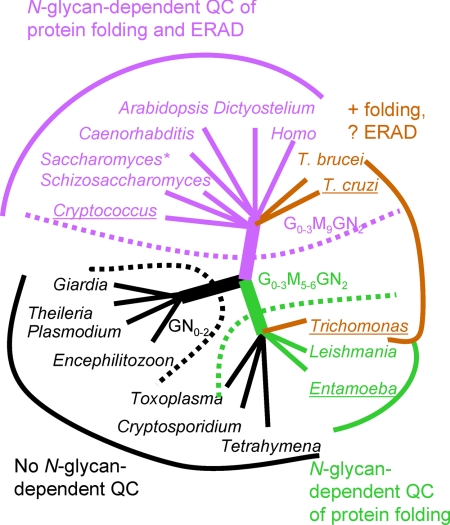
Fig. 2.
N-glycan precursors do not accurately predict the presence or absence of N-glycan-dependent QC systems for glycoprotein folding and ERAD. In this tree, organisms are grouped according to their N-glycan precursors (Table 1) (4). Encephalitozoon, Theileria, Plasmodium, and Giardia, which have N-glycan precursors composed of GN0–2, are predicted to have no N-glycan-dependent QC of glycoprotein folding and degradation (dotted black line). L. major, Trichomonas, Entamoeba, Tetrahymena, Cryptosporidium, and Toxoplasma, which have N-glycan precursors composed of Glc0–3Man5–6GlcNAc2, are predicted to have N-glycan-dependent QC of glycoprotein folding only (dotted green line). Saccharomyces, Schizosaccharomyces, Cryptococcus, Homo, Arabidopsis, Dictyostelium, T. brucei, and T. cruzi, which have N-glycan precursors composed of Glc0–3Man9GlcNAc2, are predicted to have N-glycan-dependent QC of glycoprotein folding and ERAD (dotted purple line). Results from protein predictions (Table 1), phylogenetic trees (Fig. 3), and experiments (Figs. 4–7) are shown with solid colored lines and names for each organism, where black again indicates no N-glycan-dependent QC, green indicates N-glycan-dependent QC of folding, and purple indicates N-glycan-dependent QC of folding and ERAD. Brown indicates organisms where the bioinformatic and experimental data demonstrate N-glycan-dependent QC of glycoprotein folding and suggest the possibility of N-glycan-dependent ERAD. Underlines beneath names of organisms indicate those that were included in in vitro or in vivo experiments.