Abstract
Leucocyte adhesion deficiency (LAD) is a hereditary disorder caused by mutations in the CD18 (β2 integrin) gene. Four missense mutations have been identified in three patients. CD18(A270V) supports, at a diminished level, CD11b/CD18 (Mac-1, αMβ2 integrin) and CD11c/CD18 (p150,95, αXβ2 integrin) expression and function but not CD11a/CD18 (LFA-1, αLβ2 integrin) expression. Conversely, CD18(A341P) supports a limited level of expression and function of CD11a/CD18, but not of the other two CD11/CD18 antigens. CD18(C590R) and CD18(R593C) show a decreasing capacity to associate with the CD11a, CD11c and CD11b subunits. Transfectants expressing the CD11a/CD18 with the C590R and R593C mutations are more adhesive than transfectants expressing wild-type LFA-1, and express the reporter epitope of the monoclonal antibody 24 constitutively. Thus, the four mutations affect CD18 differently in its capacities to support CD11/CD18 expression and adhesion. These results not only provide a biochemical account for the clinical diversity of patients with leucocyte adhesion deficiency, but also offer novel insights into the structural basis of interaction between the α and β subunits, which is an integral component in our understanding of integrin-mediated adhesion and its regulation.
Keywords: integrin, CD11/CD18, leucocyte adhesion deficiency
Introduction
Leucocyte adhesion deficiency (LAD) is associated with defects in the gene for the integrin β2 subunit, or, in the leucocyte antigen nomenclature, the CD18 antigen [1–4]. The defect has been attributed primarily to a diminished level of expression of the leucocyte integrins LFA-1 (CD11a/CD18 or the αLβ2 integrin), Mac-1 (CD11b/CD18 or the αMβ2 integrin) and p150,95 (CD11c/CD18 or the αXβ2 integrin) on the patients' leucocytes [3–5]. Clinically, LAD patients are characterized by the elevation of blood neutrophil counts, recurrent bacterial and fungal infections, slow wound healing and dystrophic scars from skin injuries. In most cases, the degree of severity of these symptoms may be correlated with the level of CD11/CD18 expression on the patients' leucocytes. Generally, patients with a less than 1% normal level of CD11/CD18 are susceptible to frequent and life-threatening systemic infections. Patients with higher levels of CD11/CD18 expression, up to 10% of normal, may survive to adulthood with proper medical care [4,5]. This heterogeneity of clinical phenotype is reflected by the heterogeneity of CD18 mutations. To date, there are over 20 different mutations described at the molecular level [4,6,7]. However, none of CD11/CD18 with these mutations was shown to possess any residual adhesion activities even though two of them, S138P [6] and D231H [7], were shown to be capable of supporting expression of LFA-1, Mac-1 and p150,95. Thus, there is a gap in the biochemical correlation between genotype and phenotype of LAD, particularly in the moderate cases where CD11/CD18 expression and functions are not completely abolished. In this article, we report four CD18 mutations identified in three LAD patients GF, HM and TB. We show that CD18 with these mutations have differential ability to associate with the three CD11 subunits to form the LFA-1, Mac-1 and p150,95 antigen complexes, and that the mutant CD11/CD18 integrins, when expressed, have detectable ligand binding activities.
Materials and methods
Patients GF, HM and TB
GF is a woman of Iranian origin (ethnically Persian) and was born in 1982. She suffers from chronic gingivitis, recurrent oral and vaginal ulceration with scarring, and cutaneous scalp and vaginal fungal infections. LAD was confirmed by flow cytometric analysis. For a period between July 1996 and January 1998, LFA-1 and Mac-1 levels on GF's neutrophils and lymphocytes were followed and showed a variation between 1 and 11%.
HM (1986–98) was of Mexican Hispanic background and had only ∼1% of CD11b/CD18 on her neutrophils [8]. After a match-related bone marrow transplant in October 1997, she developed severe graft-versus-host disease involving the skin. Despite medical management, she eventually developed severe pneumonitis, which led to her demise in January 1998.
TB (1963–93) was a Gypsy born in a family with LAD history [9,10]. Both LFA-1 (∼10%) and Mac-1 (∼5%) were detected on TB's neutrophils. Killer, natural killer and cytotoxic T-cell activity, helper T-cell activity and phytohaemagglutinin-induced proliferation were demonstrated with TB's lymphocytes, and these activities were more sensitive to the inhibitory effect of anti-LFA-1 MoAbs. TB died in 1993 in a car accident.
Reagents
cDNAs of CD11 and CD18 in the expression vector pcDNA3 were described previously [11]. The monoclonal antibodies were gifts from different sources: MHM23 (anti-CD11/CD18) and MHM24 (anti-CD11a) [12,13] from Professor A. J. McMichael (Institute of Molecular Medicine, Oxford, UK); LPM19c (anti-CD11b) [14] and KB43 (anti-CD11c) [7] from Dr K. Pulford (LRF Diagnostic Unit, Oxford, UK); MEM-48 (anti-CD18, activating MoAb) [15] from Professor V. Horejsi (Prague, Czech Republic); KIM185 (anti-CD18, activating MoAb) [16] from Dr M. Robinson (Celltech Therapeutics, Ltd, Slough, UK); and MoAb 24 (activation reporter MoAb for CD11a/CD18) [17] from Dr N. Hogg (ICRF, London, UK). ICAM-1/Fc, ICAM-2/Fc and ICAM-Fc were prepared as previously described [18]. iC3b was prepared according to Cai and Wright [19].
Sequence analysis of the CD18 subunit alleles
Poly A RNA was obtained from Epstein-Barr virus (EBV)-transformed cells from the three patients using the Quickprep Micro mRNA purification kit (Pharmacia Biotech, St Albans, UK). CD18 cDNA was obtained and sequenced as previously described [6]. Exon 9, in which the A341P mutation was located, was amplified from the genomic DNA of patient HM using the oligonucleotides I89F and I910R. The C534* mutation was located to exon 12 by PCR sequencing from the genomic DNA of HM. Exon 7 (A270V) was amplified from the genomic DNA of patient GF using the oligonucleotide pair E94/03 and E94/04, and exon 13 (C590R) with I1213F and I1314R. The fragments were cloned into PCR-Script Amp SK(+) vector (Stratagene Ltd, Cambridge, UK) and sequenced. The defect in the CD18 genes of the patient TB was identified by PCR sequencing of the CD18 cDNA. A single sequence with the mutation R593C was found in exon 13 of PCR amplified genomic DNA.
The oligonucleotides E94/03 and E94/04 were described previously [6]. The sequences of the other oligonucleotides are as follows: I89F, AGCCGTGCCATGTCTGGTC; I910R, TGAGTG GTGCGGGAGACTC; I1213F, AAGCAGGAGCTTAGCCGTG; and I1314R, TGCTCCGCCTGCACTCAC.
Transfection and adhesion assays
Appropriate cDNA fragments containing each of the four mutations were spliced into a wild-type CD18 cDNA clone [20] in the expression vector pcDNA3 (Invitrogen BV, Groningen, The Netherlands). CD11 cDNA (5 µg) and 1 µg of CD18 cDNA were transfected into COS-7 cells in 80 cm2 flasks using the DEAE-dextran method [20] as previously described [11,21,22]. The MoAb MHM23, which is specific for CD11/CD18 heterodimers, gives different flow cytometric backgrounds on COS-7 cells transfected with the CD11 cDNA alone. Thus, expression of the heterodimers on CD11/CD18 transfectants was measured over the background of the respective CD11 transfectants. Subtractive histogram analyses were performed using the CellQuest software (Becton Dickinson, Oxford, UK).
Microtitre wells were coated with ICAM-Fc, iC3b or denatured BSA as described [6,22]. COS-7 transfectants were resuspended in RPMI-1640 with 5% fetal calf serum and 10 mm HEPES, pH 7·5 (RPMI-FH), to between 0·5 and 1 × 106 cells per ml. The fluorescent dye BCECF-AM (Molecular Probes, Leiden, The Netherlands), at 1 µg/µl in DMSO, was added to a final concentration of 1 µg/ml, and labelling was carried out at 37°C for 20 min. The cells were washed twice and resuspended to yield 2–4 × 105 cells per ml. A 50 µl volume of the cell suspension was dispensed into each well of the ligand-coated microtitre plates. The plates were incubated at 37°C for 30 min. The total number of cells in each well was quantified by measuring the fluorescence signal using the Cytofluor 4000 fluorescent plate reader (PE Biosystems, Warrington, UK). Percentage of cell adhesion was determined as the relative fluorescence signal after washing three times with 150 µl RPMI-FH.
CD11/CD18 mediated adhesion to the various ligands was activated with the MoAbs MEM48 (10 µg/ml) or KIM185 (2·5 µg/ml). The blocking MoAbs used were MHM24 (10 µg/ml) for CD11a/CD18 mediated adhesion, LPM19c (10 µg/ml) for CD11b/CD18 mediated adhesion, and KB43 (10 µg/ml) for CD11c/CD18 mediated adhesion, respectively. The concentrations of the activating and blocking antibodies used were pre-titrated to give maximal adhesion and complete blocking.
Expression of the activation reporter epitope of MoAb 24 on LFA-1 transfectants was monitored by flow cytometry. The cells were stained with MoAb 24, at 20 µg/ml, in RPMI-FH media, or in HEPES BSA buffer (10 mm HEPES, 150 mm NaCl, 0·1% BSA) with 5 mm MgCl2 and 1·5 mm EGTA (Mg/EGTA) at 37°C for 30 min. The cells were washed twice in the same buffer before staining with the FITC-conjugated second antibody in RPMI-FH media.
Results
Molecular characterization of defects
Poly A RNA was obtained from EBV-transformed cell lines of the three patients. CD18 cDNA fragments were obtained by RT-PCR as previously described [6]. These fragments were sequenced and the results are summarized in Table 1. Two mutations, A270V and C590R, were found in the cDNA of patient GF. The A270V mutation is associated with the abolition of the restriction enzyme site BstUI. Partial digestion with BstUI was obtained for the cDNA as well as for the genomic fragment spanning exon 7 (data not shown). The mutation A341P was found in 10 out of 10 CD18 clones from patient HM. This mutation is associated with the creation of a Tsp45I restriction site. Whereas the cDNA fragment was completely digested with Tsp45I, only partial digestion was obtained for the genomic fragment (data not shown). This result suggested that the other CD18 allele of HM was not expressed at the RNA level. PCR sequencing of the genomic DNA of HM revealed a double sequence in exon 12. One sequence was normal and the other contained a C to A mutation causing the conversion of the TGC codon for the cysteine at position 534 to the stop codon TGA. This mutation will be referred to as C534*. Thus, both GF and HM are compound heterozygotes for LAD. Patient TB was shown to be homozygous for the mutation R593C by PCR sequencing of his genomic DNA.
Table 1.
Mutations in the CD18 alleles of patients GF, HM and TB
| Nucleotide change † | cDNA | Genomic DNA | ||||||
|---|---|---|---|---|---|---|---|---|
| Patients | Mutations * | Normal | Variant | Enzyme site change | location ‡ | # clones variant/total | exon location § | # clones variant/total |
| GF | A270V | C(GCG) | C(GTG) | BstUI lost | 809 | 12/17 | 7 | 3/5 |
| C590R | (TGT) | (CGT) | none | 1768 | 9/22 | 13 | 3/4 | |
| HM | A341P | GTCA(GCC) | GTCA(CCC) | Tsp45I created | 1021 | 10/10 | 9 | 3/6 |
| C534* | C(TGC)G | C(TGA)G | DdeI created | 1602 | – | 12 | – | |
| TB | R593C | (CGT) | (TGT) | none | 1777 | 3/3 | 13 | – |
Mutations in the CD18 protein sequence. The initiation methionine is assigned number ‘1’.
Mutations in the cDNA sequence. For clarity, the DNA sequences and numbering are shown in italics. The altered bases are shown in bold and the codons involved are in brackets. Where restriction enzyme sites are affected, the sequences are shown and underlined.
The location of the altered nucleotides in the cDNA sequence. The ‘A’ in the initiation codon ‘ATG’ is assigned number ‘1’.
Exon locations are assigned according to Weitzman et al. [20].
A270V, A341P and C590R are novel mutations found in the CD18 gene of LAD patients. The C543* and R593C mutations have been previously reported. A LAD patient of the severe phenotype was found to be homozygous for the C534* mutation [23]. The R593C mutation has been reported in two patients of the moderate phenotype. Both patients were compound heterozygotes: one patient carried the K196T and R593C alleles [24], and the other patient the G284S and R593C alleles [25].
Functional analyses
The four missense mutations were each separately introduced into the CD18 cDNA in the expression vector pcDNA3. Each of them was transfected into COS-7 cells together with the cDNA, also in pcDNA3, of either CD11a, CD11b or CD11c. The transfectants were analysed for the expression of the CD11/CD18 heterodimers and their ability to mediate specific adhesion to a panel of ligands, ICAM-1, -2 and -3 for LFA-1 (CD11a/CD18) [26], and iC3b [27,28] and denatured BSA [29] for the Mac-1 (CD11b/CD18) and p150,95 (CD11c/CD18) antigens.
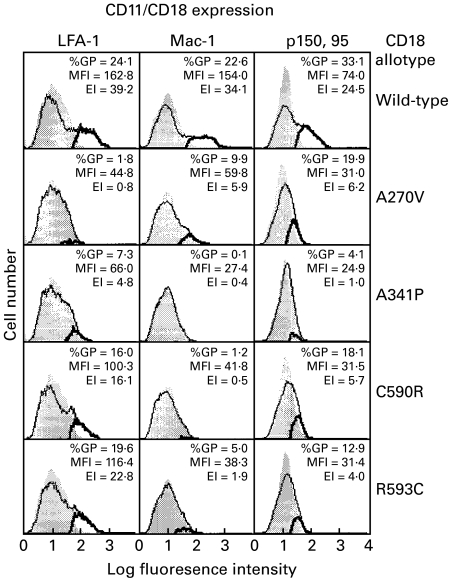
Heterodimer expression was assessed by flow cytometry with the dimer-specific MoAb MHM23 [12,13,22]. Since we were trying to observe subnormal levels of expression of the mutants, we employed the subtractive histogram analysis in the CellQuest software. The expression profile of the MHM23 epitope in the double transfectants was compared with those seen in the CD11 transfectants (Fig. 1). None of the CD18 mutants supported full CD11/CD18 expression, although the expression index of CD11a/CD18(R593C) was over 50% that of the wild type. The most intriguing observation was that the four mutants showed different capacities to combine with the three CD11 subunits. A270V combined more efficiently with CD11c than with CD11b, and least efficiently with CD11a. The other three mutants, namely A341P, C590R and R593C, showed a preference to combine with CD11a followed by CD11c and CD11b. Similar results were obtained with another CD11/CD18 heterodimer specific MoAb, IB4 [30].
Fig. 1.
Expression of wild-type and mutant CD11/CD18 antigens on COS-7 transfectants. Wild-type CD18 cDNA or CD18 cDNA with the A270V, A341P, C590R and R593C mutations were transfected into COS-7 cells together with the wild-type cDNA of CD11a, CD11b or CD11c. CD11/CD18 expression was monitored by flow cytometry with the MoAb MHM23. The background histograms (shadow) are those obtained with transfectants of CD11a, CD11b or CD11c alone. Expression profiles of the CD11/CD18 transfectants are shown in solid lines. The subtracted profiles, using the CellQuest software, are shown in bold lines. Percentage of gated positive cells (%GP) in the subtracted profiles, the mean fluorescence intensity (MFI) and the expression index (EI) = %GP × MFI are also shown.
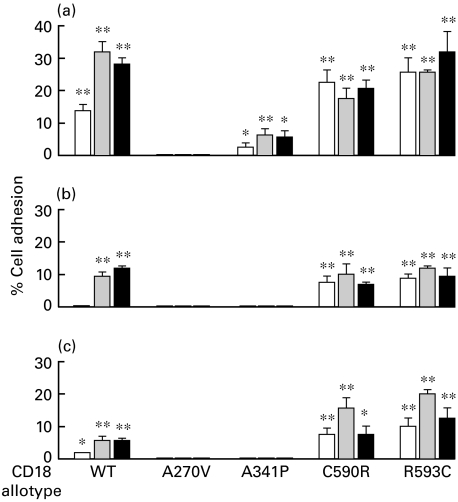
The adhesion of wild-type LFA-1 transfectants to ICAM-1 can be detected in the absence of stimulation, and the level of adhesion can be promoted to a higher level with either activating MoAb MEM48 [15] or KIM185 [16] (Fig. 2). Adhesion to ICAM-2 and ICAM-3 is uniformly lower, and it can only be detected in the presence of the activating MoAbs. Transfectants expressing the CD11a/CD18(A341P) have a similar, but reduced, adhesion profile to ICAM-1, and their adhesion to ICAM-2 and ICAM-3 is not detected. In contrast, both CD11a/CD18(C590R) and CD11a/CD18(R593C) transfectants are more adherent than the wild-type, as higher levels of adhesion are consistently demonstrated with the variants in the absence of the activating MoAbs, though the level of CD11a/CD18 expression is always lower than the wild type (see Fig. 1). Their adhesion to ICAM-3, as shown in Fig. 2, can be further promoted with MEM-48. To summarize, these results show that CD18(A270V) supports LFA-1 formation poorly, CD18(A341P) supports a reduced but detectable level of LFA-1 formation and adhesion, and LFA-1 with the CD18(C590R) and CD18(R593C) are more adhesive than the wild type.
Fig. 2.
Adhesion of LFA-1 (CD11a/CD18) transfectants to (a) ICAM-1, (b) ICAM-2 and (c) ICAM-3. Adhesion was done in the absence (□) or the presence of the activating MoAbs (▪) MEM-48 (10 µg/ml) or (▪) KIM185 (2·5 µg/ml). The data shown are after subtraction of background obtained in the presence of the blocking MoAb MHM24 (10 µg/ml). Adhesion in the absence and presence of the blocking MoAb were compared by one-tail Studentss t-test assuming unequal variance: *, P < 0·05; **, P < 0·01.
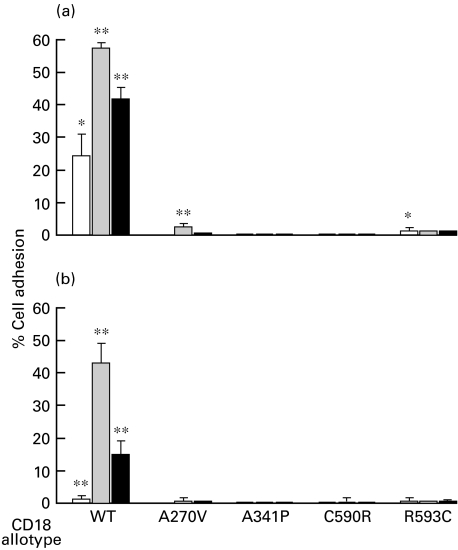
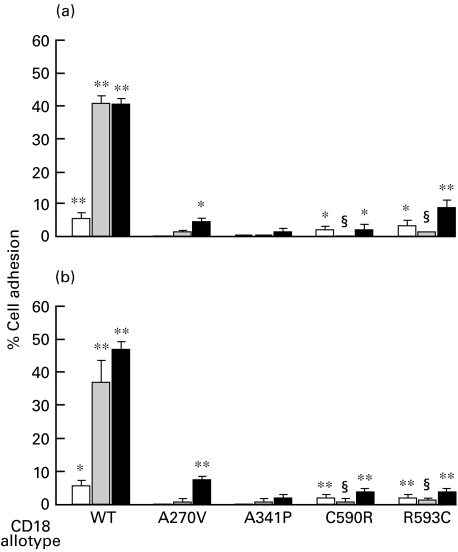
Low levels of adhesion of wild-type Mac-1 and p150,95 transfectants to iC3b and denatured BSA were detected in the absence of stimulation. These levels could be enhanced with the activating MoAbs MEM48 or KIM185 (Figs 3, 4). Because of the low level of expression, adhesion of Mac-1 with the CD18 mutations to iC3b and denatured BSA is difficult to detect (Fig. 3). Adhesion of CD11c/CD18(A270V) to iC3b and denatured BSA can be detected in the presence of KIM185 (Fig. 4). Adhesion of CD11c/CD18(C590R) and CD11c/CD18(R593C) mutants to iC3b and denatured BSA can be detected in the absence of the activating MoAbs (Fig. 4). Curiously, we consistently observe a reduction in adhesion of these two variants in the presence of MEM-48, which has the opposite effect on the wild-type CD11c/CD18.
Fig. 3.
Adhesion of Mac-1 (CD11b/CD18) transfectants to (a) iC3b and (b) denatured BSA. Adhesion was done in the absence (□) or the presence of the activating MoAbs (▪) MEM-48 (10 µg/ml) or (▪) KIM185 (2·5 µg/ml). The data shown are after subtraction of background obtained in the presence of the blocking MoAb LPM19c (10 µg/ml). Adhesion values in the absence and presence of the blocking MoAb were compared by one-tail Student's t-test assuming unequal variance: *, P < 0·05; **, P < 0·01.
Fig. 4.
Adhesion of p150,95 (CD11c/CD18) transfectants to (a) iC3b and (b) denatured BSA. Adhesion was done in the absence (□) or the presence of the activating MoAbs (▪) MEM-48 (10 µg/ml) or (▪) KIM185 (2·5 µg/ml). The data shown are after subtraction of background obtained in the presence of the blocking MoAb KB43 (10 µg/ml). Adhesion values in the absence and presence of the blocking MoAb were compared by one-tail Student's t-test assuming unequal variance: *, P < 0·05; **, P < 0·01. Adhesion values of CD11c/CD18 with the C590R and R593C mutations in the presence of MEM-48 were also compared with the values obtained in the absence of MEM-48 with Student's t-test: §, P < 0·05.
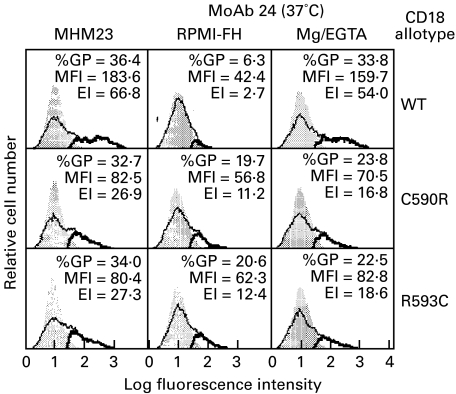
Adhesion of CD11a/CD18 to ICAM-1 is associated with the expression of the reporter epitope of MoAb 24 [17]. Thus, whereas the expression of the MoAb 24 epitope on CD11a/CD18 transfectants is low in RPMI-FH media, its expression can be enhanced with Mg/EGTA. In contrast, the epitope of MoAb 24 is detected at significant levels without stimulation on transfectants expressing the CD11a/CD18 variants with the C590R and R593C mutations (Fig. 5).
Fig. 5.
Expression of the activation reporter epitope of MoAb 24 on CD11a/CD18 transfectants. Expression of the CD11a/CD18 heterodimers was monitored with the MoAb MHM23. Detection of the MoAb 24 epitope was performed at 37°C in RPMI-FH medium or in HEPES BSA buffer supplemented with 5 mm MgCl2 and 1·5 mm EGTA (Mg/EGTA). %GP, MFI and EI are as in Fig. 1.
Discussion
Like the clinical features, the molecular nature of the CD18 defects among LAD patients is highly heterogeneous [2–5]. However, there is limited biochemical information to correlate a particular mutation to a particular set of clinical symptoms. Generally, mutations that lead to grossly abnormal gene products may be associated with the severe LAD phenotype [3–5]. These mutations include nucleotide deletions leading to frameshifts (three cases), abnormal splicing defects (four cases), a point mutation leading to a defective initiation codon (one case), and a nonsense mutation (one case). Other defects are missense mutations, in which a single amino acid is changed in the CD18 protein. In these cases, the biochemical consequences are more subtle, and how each mutation affects the expression and function of the CD11/CD18 antigens has to be assessed individually. We report here the study of three LAD patients. Patient GF is a compound heterozygote with the A270V and C590R missense mutations. Patient HM was also a compound heterozygote but only the A341P allele was expressed at the mRNA level. The other mutation is a nonsense mutation with the TGC codon for C534 converted into the stop codon TGA (C534*). The C534* mutation was previously reported to have low expression at the mRNA level [23]. It is a common finding that the presence of nonsense mutations reduces the steady state level of the mRNA (see the Discussion in [23]). Patient TB was homozygous for the mutation R593C.
The COS-7 cell expression system is designed to over-express the proteins coded for by the transfected DNA. For this reason, this system provides us with the advantage to examine the level of the expressed proteins under magnification and therefore, to distinguish between the mutations that do not support expression from those that do so at a low level. It has been used by Wardlaw et al. [31] to demonstrate that whereas the CD18 mutation L149P can support low levels of CD11a/CD18 expression, CD18 with the G169R mutation cannot. In our laboratory, we have shown that mutations G273R [6] and G284S [7] are incapable of supporting the CD18 expression in combination with CD11a, CD11b and CD11c. Conversely, significant expression of the three CD11/CD18 antigens was found with the mutants S138P [6] and D231H [7], although transfectants expressing these variants were not adherent to ligands. Using this expression system, all four missense mutations reported here are found to support a reduced level, to varying degrees, of CD11/CD18 expression. Furthermore, their capacity to support CD11/CD18 surface expression is not uniform with the three CD11 subunits. Thus, CD18(A270V) shows preference for CD11c, followed by CD11b then CD11a. In contrast, CD18(A341P), CD18(C590R) and CD18(R593C) show a descending order of preference for CD11a, CD11c and CD11b (Fig. 1).
The extracellular domain of the CD18 antigen is organized, according to the primary structure [32,33], into four regions: an N-terminal cysteine-rich region which may have a PSI (plexin, semaphorin and integrin) domain [34], a highly conserved region (HCR), which, by structural prediction, may assume an I-domain-like fold [35–37], and a mid-region which links the HCR to the major cysteine-rich region (CRR), in which 39 cysteines are arranged in a conserved and characteristic pattern [32,33]. To date, 14 CD18 missense mutations, including the four reported here, have been identified in LAD patients; 12 of these are found in the HCR and the other two in the major CRR.
The A270V and A341P mutations are located in the HCR. They are distinct from those that do not support CD11/CD18 expression, such as G169R [31], G273R [6] and G284S [7], and those that support expression but not adhesion, such as S138P [6] and D231H [7]. Low levels of CD11c/CD18 expression can be detected with the A270V mutant, and CD11a/CD18 expression with A341P. In both cases, COS-7 transfectants expressing the mutant integrins can be stimulated to bind ligands like their wild-type counterparts. The levels of adhesion are lower in reflection of their lower CD11/CD18 expression levels. These results show that the different residues in the HCR of CD18 can affect its efficiency in forming the different CD11/CD18 complexes.
The C590R and R593C are the only two missense mutations located outside the HCR. The CRR of the integrin β subunits is intriguing not only because of the high number of cysteine residues in the region, but also the very conserved and characteristic pattern in which they are arranged. It is thus anticipated that the mutation of a cysteine residue in this region, such as C590R, or the introduction of an extra one, such as R593C, would affect the normal disulphide bonding pattern in this region. It is therefore unexpected to find that the expression indices for LFA-1 variants with the C590R and R593C mutations are 41% and 58%, respectively, that of the wild type (see Figs 1, 5). CD18 with the C590R or R593C mutations is less effective in supporting the expression of CD11c/CD18 and, to an even lesser extent, of CD11b/CD18. Previously, we have described a chimeric CD18/CD29 molecule (β2/β1 integrin subunit), in which the CRR of CD18 was replaced with that of CD29 [21], and its capacity to support CD11a/CD18 expression on COS-7 transfectants. Subsequent work showed that it is less effective in supporting Mac-1 and p150,95 expression, though the presence of the two integrins are easily detected on the cell surface (R. H. Hyland and S. K. A. Law, unpublished observation). These findings are consistent with the two CRR missense mutations reported here and, together, they indicate that the CRR is more important for Mac-1 and p150,95 heterodimer expression than for LFA-1.
LFA-1 variants with the CRR mutations bind ICAM-1 constitutively, and the addition of the activating MoAbs MEM-48 and KIM185 has no effect (Fig. 2). Their adhesion to ICAM-2 and ICAM-3 is significantly higher than the wild-type LFA-1 transfectants, and, in the case of ICAM-3, adhesion can be further promoted with the MoAb MEM-48 (Fig. 2). Expression of the MoAb 24 epitope can be induced on wild-type LFA-1 by Mg/EGTA at 37°C, and it has been associated with an LFA-1 conformation that is adhesive to ICAM-1 [17]. Since the MoAb 24 epitope is expressed on the LFA-1 variants with the CRR mutations without the manipulation of the divalent cations (Fig. 5), we may conclude that the variants assume an active conformation with respect to ICAM-1 adhesion. This finding is reminiscent of our observations on the CD18/CD29 chimeric integrin subunits; LFA-1 with the CD29 CRR is constitutively active with respect to ICAM-1 adhesion, and expresses the MoAb 24 epitope in the absence of Mg/EGTA [21]. Thus, similar functional effects are observed with the exchange of the CD18 CRR with that of CD29 and with the introduction of either the C590R or the R593C mutations. Since the two mutations involve cysteine residues, it is reasonable to speculate that perturbation of the disulphide bond pattern of the CRR is the primary cause leading to the observed consequences. However, how they affect the disulphide bonds in the CRR is unclear. A disulphide bond model for the β3 integrin subunit (CD61) has been proposed [38] but, because of the CRR's extreme resistance to proteolysis, the disulphide bonds in the CRR were assigned based on the arguments that there are four pseudo-repeating elements in the CRR, and that the disulphide bonds within each repeating unit are self-contained. Thus, the assignments, though reasonable, are not supported by biochemical data and are therefore difficult to use as the basis for extended analysis. (For detailed discussion, see [39].)
It should be noted that CD11c/CD18 with the two CRR mutations are expressed at sufficient levels on the transfectants, and low levels of adhesion to iC3b and BSA can be detected. Curiously, MEM-48 is inhibitory to the adhesion of the CD11c/CD18 CRR variants but it activates the adhesion of the wild type (Fig. 4). Furthermore, the inhibitory effect is not due to the mutations themselves, since MEM-48 promotes adhesion to ICAM-3 of CD11a/CD18 with the same mutations (Fig. 2). Detailed studies of the biochemical properties of the CD11/CD18 integrins with the CRR mutations, and their response to activating and inhibitory MoAbs, will be required to further understand the regulatory mechanism of the CD11/CD18 integrins.
A summary of the comparative surface expression of the CD11/CD18 integrins on the leucocytes of the three LAD patients and COS transfectants is shown in Table 2. Although the information on the patients' leucocytes is not complete, the surface expression of a particular CD11/CD18 integrin, when detected on a patient's leucocytes, generally correlates with its expression on COS transfectants with one notable exception. The CD18 with either the C590R or R593C mutation supports CD11a/CD18 expression on COS transfectants, but the antigen was detected at a significantly lower level on the leucocytes of GF and TB. The CD11a/CD18 variants on COS transfectants are constitutively active with respect to ICAM-1 adhesion and express the MoAb 24 epitope. We hypothesize that there may exist mechanisms specific in leucocytes to down-regulate the expression of active integrins, to be overcome when the leucocytes are stimulated to become adherent. Thus, although the CD11a/CD18 antigens are only found at low levels on the patient's leucocytes, they may be stimulated, as in the case of TB's T cells upon culturing in phytohaemagglutinin, to express up to 50% normal level of CD11a/CD18 [9]. These mechanisms are absent in COS cells and the CD11a/CD18 antigen is expressed irrespective of its activation state. Future experimental work is required to test the validity of this hypothesis.
Table 2.
Summary of CD11/CD18 expression on leucocytes of LAD patients and COS transfectants
| CD11/CD18 expression (% of normal) | |||||
|---|---|---|---|---|---|
| Patients and mutations * | Cells | CD18 | CD11a | CD11b | CD11c |
| GF (A270V/C590R) | Neutrophils | 2–8 | 2–11 | 0–7 | 45 |
| Monocytes | 0–2 | 0–4 | 0–6 | 12 | |
| Lymphocytes | 1–2 | 0–3 | NA † | NA | |
| COS (A270V)‡ | 2 | 17 | 25 (R)§ | ||
| COS (C590R) | 41 (CA)¶ | 1 | 23 (R) | ||
| HM (A341P/C534*) | Neutrophils | NA | NA | 1 | NA |
| Monocytes | NA | NA | NA | NA | |
| Lymphocytes | NA | NA | NA | NA | |
| COS (A341P) | 12 (R) | 1 | 4 | ||
| TB (R593C) | Neutrophils | 11–12 | 2–9 | 2–7 | 21 |
| Monocytes | 0 | 0 | 0 | 0 | |
| Lymphocytes | 0 | 0 | NA | NA | |
| COS (R593C) | 41 (CA) | 6 | 16 (R) | ||
CD11/CD18 expression data on GF's leucocytes were collected and analysed aperiodically since 1994. Only CD11b/CD18 expression data on HM's neutrophils were available [8]. The data on TB were from [10]. A270V and A341P mutations are located in the HCR and the C590R and R593C mutations in the CRR. CD18 with the C534* is not detected at the mRNA level.
NA: not available.
COS cell expression values were obtained from the percentage value of the expression indices (Fig. 1) of the mutant transfectants with reference to the wildtype transfectants.
R: residual ligand adhesion activities detected.
The results obtained from laboratory analyses can also be correlated, in broad terms, with the clinical features of the LAD patients, in particular with GF and TB. GF has a mild LAD phenotype, and her infections are generally kept under control with antibiotics. It may be rationalized that her two CD18 mutant alleles, C590R and A270V, support minimal expression of functional CD11a/CD18 and CD11c/CD18, respectively, and collectively provide her with a limited scope of CD11/CD18 mediated activities. TB had a moderate LAD phenotype. His T cells can be stimulated to express 50% of normal level of CD11a/CD18. In addition, many lymphocyte functions can be detected with TB's cells and they were inhibited by MoAbs against CD11a/CD18 [9,10], suggesting that the CD11a/CD18 expressed on his lymphocytes was functional. This is consistent with the laboratory finding that the R593C mutation can support CD11a/CD18 expression and adhesion, though qualitatively different from that of the wild type. HM's neutrophils expressed CD11b/CD18 at about 1% that of normal, which is reflective of their diminished capacity to adhere to fibrinogen, vitronectin and laminin [8]. This observation is consistent with the CD18 mutation A341P which showed minimal support of CD11b/CD18 expression (Fig. 1). Unfortunately, the full CD11/CD18 expression profile of HM's leucocytes was not obtained when available.
We have shown that mutations in both the HCR and major CRR can affect CD18 in its ability to support CD11/CD18 expression and function, and that the different CD18 mutants interact differently with the CD11a, CD11b and CD11c subunits. These results have provided the biochemical link necessary to correlate the different mutations to the diverse clinical phenotypes among the LAD patients, and insights into the contribution of the HCR and the major CRR in heterodimer expression and functions of the CD11/CD18 integrins.
Acknowledgments
This work was supported in part by grants from the Arthritis Research Campaign (UK), the National Institute of Health (Bethesda, MD), The Wellcome Trust (UK) and the European BIOMED 2 Concerted Action. We thank R. H. Hyland and E. C. Mathew for comments on the manuscript.
References
- 1.Crowley CA, Curnutte JT, Rosin RE, et al. An inherited abnormality of neutrophil adhesion: its genetic transmission and its association with a missing protein. New Engl J Med. 1980;302:1163–8. doi: 10.1056/NEJM198005223022102. [DOI] [PubMed] [Google Scholar]
- 2.Springer TA, Thompson WS, Miller LJ, Schmalsteig FC, Anderson DC. Inherited deficiency of the Mac-1, LFA-1, p150,95 glycoprotein family and its molecular basis. J Exp Med. 1984;160:1901–18. doi: 10.1084/jem.160.6.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson DC, Schmalsteig FC, Finegold MJ, et al. The severe and moderate phenotypes of heritable Mac-1, LFA-1 deficiency: their quantitative definition and relation to leukocyte dysfunction and clinical features. J Infect Dis. 1985;152:668–89. doi: 10.1093/infdis/152.4.668. [DOI] [PubMed] [Google Scholar]
- 4.Anderson DC, Kishimoto TK, Smith CW. Leukocyte adhesion deficiency and other disorders of leukocyte adherence and motility. In: Shriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic basis of inherited diseases. 7. New York: MacGraw-Hill Inc; 1994. pp. 3955–94. [Google Scholar]
- 5.Arnaout MA. Leukocyte adhesion molecules deficiency: its structural basis, pathophysiology and implications for modulating the inflammatory response. Immunol Rev. 1990;114:145–80. doi: 10.1111/j.1600-065x.1990.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 6.Hogg N, Stewart MP, Scarth SL, et al. A novel leukocyte adhesion deficiency caused by expressed but nonfunctional β2 integrins Mac-1 and LFA-1. J Clin Invest. 1999;103:97–106. doi: 10.1172/JCI3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathew EC, Shaw JM, Bonilla FA, Law SKA, Wright DW. A novel point mutation in CD18 causing the expression of dysfunctional CD11/CD18 leukocyte integrins in a patient with leukocyte adhesion deficiency (LAD) Clin Exp Immunol. 2000;121:133–8. doi: 10.1046/j.1365-2249.2000.01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suchard SJ, Burton MJ, Dixit VM, Boxer LA. Human neutrophil adherence to thrombospondin occurs through a CD11/CD18-independent mechanism. J Immunol. 1991;146:3945–52. [PubMed] [Google Scholar]
- 9.Miedema F, Tetteroo PAT, Terpstra FG, et al. Immunologic studies with LFA-1- and Mo1-deficient lymphocytes from a patient with recurrent bacterial infections. J Immunol. 1985;134:3075–81. [PubMed] [Google Scholar]
- 10.Berkinshaw CJ, Weemaes CMR, Roos D, Tetteroo PAT, Weening RS. Congenital deficiency of leukocyte-adherence glycoproteins: a familial defect. Neth J Med. 1987;31:158–70. [PubMed] [Google Scholar]
- 11.Al-Shamkhani A, Law SKA. Expression of the H52 epitope on the β2 subunit is dependent on its interaction with the α subunits of the leukocyte integrins LFA-1, Mac-1 and p150,95 and the presence of Ca2+ Eur J Immunol. 1998;28:3291–300. doi: 10.1002/(SICI)1521-4141(199810)28:10<3291::AID-IMMU3291>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 12.Hildreth JEK, Gotch FM, Hildreth PDK, McMichael AJ. A human lymphocyte-associated antigen involved in cell-mediated lympholysis. Eur J Immunol. 1983;12:202–8. doi: 10.1002/eji.1830130305. [DOI] [PubMed] [Google Scholar]
- 13.Hildreth JEK, August JT. The human lymphocyte function-associated (HLFA) antigen and a related macrophage differentiation antigen (HMac-1): functional effects of subunit-specific monoclonal antibodies. J Immunol. 1985;134:3272–80. [PubMed] [Google Scholar]
- 14.Uciechowski P, von Schmidt RE. Cluster report: CD11. In: Knapp WD, Dörken B, Gilks WR, et al., editors. Leucocyte typing IV white cell differentiation antigens. Oxford: Oxford University Press; 1989. pp. 543–51. [Google Scholar]
- 15.Bazil V, Stefanova I, Hilgert I, Kristofova H, Vanck S, Horejsi V. Monoclonal antibodies against human leukocyte antigens: IV, antibodies against subunits of the LFA-1 (CD11a/CD18) leukocyte adhesion glycoproteins. Folia Biol Praha. 1990;36:41–50. [PubMed] [Google Scholar]
- 16.Robinson MK, Andrew D, Rosen H, et al. Antibody against the Leu-CAM β-chain (CD18) promotes both LFA-1- and CR3- dependent adhesion events. J Immunol. 1992;148:1080–5. [PubMed] [Google Scholar]
- 17.Dransfield I, Cabanas C, Craig A, Hogg N. Divalent cation regulation of the function of the leukocyte integrin LFA-1. J Cell Biol. 1992;116:219–26. doi: 10.1083/jcb.116.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmons DL. Cloning cell surface molecules by transient expression in mammalian cells. In: Hartley D, editor. Cellular interactions in development. Oxford: Oxford University Press; 1993. pp. 93–128. [Google Scholar]
- 19.Cai TQ, Wright SD. Energetics of leukocyte integrin activation. J Biol Chem. 1995;270:14358–65. doi: 10.1074/jbc.270.24.14358. [DOI] [PubMed] [Google Scholar]
- 20.Weitzman JB, Wells CE, Wright AH, Clark PA, Law SKA. The gene organisation of the human β2 integrin subunit (CD18) FEBS Lett. 1998;294:97–103. doi: 10.1016/0014-5793(91)81351-8. [DOI] [PubMed] [Google Scholar]
- 21.Douglass WA, Hyland RH, Buckley CD, et al. The role of the cysteine-rich region of the β2 integrin subunit in the leukocyte function-associated antigen-1 (LFA-1, αLβ2, CD11a/CD18) heterodimer formation and ligand binding. FEBS Lett. 1998;440:414–8. doi: 10.1016/s0014-5793(98)01498-7. 10.1016/s0014-5793(98)01498-7. [DOI] [PubMed] [Google Scholar]
- 22.Tan SM, Hyland RH, Al-Shamkhani A, Douglass WA, Shaw JM, Law SKA. Effect of integrin β2 subunit truncations on LFA-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) assembly, surface expression and function. J Immunol. 2000;165:2574–81. doi: 10.4049/jimmunol.165.5.2574. [DOI] [PubMed] [Google Scholar]
- 23.Lopez Rodriguez C, Nueda A, Grospierre B, et al. Characterization of two new CD18 alleles causing severe leukocyte adhesion deficiency. Eur J Immunol. 1993;23:2792–8. doi: 10.1002/eji.1830231111. [DOI] [PubMed] [Google Scholar]
- 24.Arnaout MA, Dana N, Gupta SK, Tenen DG, Fathallah DM. Point mutations impairing cell surface expression of the common β subunit (CD18) in a patient with leukocyte adhesion molecule (Leu-CAM) deficiency. J Clin Invest. 1990;85:977–81. doi: 10.1172/JCI114529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright AH, Douglass WA, Taylor GM, et al. Molecular characterization of leukocyte adhesion deficiency in six patients. Eur J Immunol. 1995;25:717–22. doi: 10.1002/eji.1830250313. [DOI] [PubMed] [Google Scholar]
- 26.Binnerts ME, van Kooyk Y. How LFA-1 binds to different ligands. Immunol Today. 1999;20:240–5. doi: 10.1016/s0167-5699(99)01467-x. 10.1016/s0167-5699(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 27.Beller DI, Springer TA, Schreiber RD. Anti-Mac-1 selectively inhibits the mouse and human type three complement receptor. J Exp Med. 1982;156:1000–9. doi: 10.1084/jem.156.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myones BL, Danzell JG, Hogg N, Ross GD. Neutrophil and monocyte cell surface p150,95 has iC3b–receptor (CR4) activity resembling CR3. J Clin Invest. 1988;82:640–51. doi: 10.1172/JCI113643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis GE. The Mac-1 and p150,95 β2 integrins bind denatured proteins to mediate leukocyte cell–substrate adhesion. Exp Cell Res. 1992;200:242–52. doi: 10.1016/0014-4827(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 30.Wright SD, Rao PE, van Voorhis WC, et al. Identification of the C3bi receptor of human monocytes and macrophages by using monoclonal antibodies. Proc Natl Acad Sci USA. 1983;80:5699–703. doi: 10.1073/pnas.80.18.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wardlaw AJ, Hibbs ML, Stacker SA, Springer TA. Distinct mutations in two patients with leukocyte adhesion deficiency and their functional correlates. J Exp Med. 1990;172:335–45. doi: 10.1084/jem.172.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishimoto TK, O'Connor K, Lee A, Roberts TM, Springer TA. Cloning of the β subunit of the leukocyte adhesion proteins: homology to an extracellular matrix receptor defines a novel supergene family. Cell. 1987;48:681–90. doi: 10.1016/0092-8674(87)90246-7. [DOI] [PubMed] [Google Scholar]
- 33.Law SKA, Gagnon J, Hildreth JEK, Wells CE, Willis AC, Wong AJ. The primary structure of the β-subunit of the cell surface adhesion glycoproteins LFA-1, CR3 and p150,95 and its relationship to the fibronectin receptor. EMBO J. 1987;6:915–9. doi: 10.1002/j.1460-2075.1987.tb04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bork P, Doerks T, Springer TA, Snel B. Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci. 1999;24:261–3. doi: 10.1016/s0968-0004(99)01416-4. [DOI] [PubMed] [Google Scholar]
- 35.Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–8. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 36.Tuckwell DS, Humphries MJ. A structure prediction for the ligand-binding region of the integrin β subunit: evidence for the presence of a von Willebrand factor A domain. FEBS Lett. 1997;400:297–303. doi: 10.1016/s0014-5793(96)01368-3. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Zang Q, Takagi J, Springer TA. Structural and functional studies with antibodies to the integrin β2 subunit: a model for the I-like domain. J Biol Chem. 2000;275:21514–24. doi: 10.1074/jbc.M002286200. [DOI] [PubMed] [Google Scholar]
- 38.Calvete JJ, Henschen A, Gonzalez-Rodriguez J. Assignment of disulphide bonds in human platelet GPIIIa: a disulphide pattern for the β-subunits of the integrin family. Biochem J. 1991;274:63–71. doi: 10.1042/bj2740063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan SM, Mathew EC, Scarth SL, Al-Shamkhani A, Law SKA. The effects of cysteine to alanine mutations of CD18 on the expression and adhesion of the CD11/CD18 integrins. FEBS Lett. 2000;486:89–92. doi: 10.1016/s0014-5793(00)02247-x. [DOI] [PubMed] [Google Scholar]