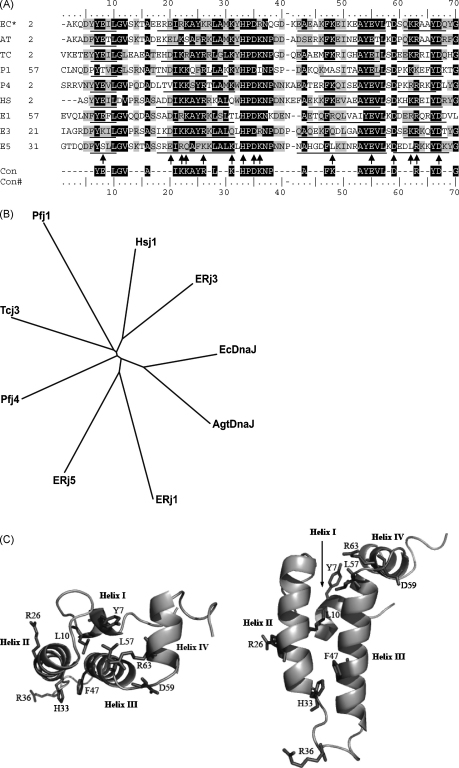
Fig. 2.
J-domains and conserved J-domain residues targeted for analysis. (A) Structure aided alignment of J-domains analyzed in this study. Predicted α-helices are underlined. Highly conserved charged residues are indicated by arrows. EC—E. coli DnaJ [GenBank accession number: P31689] residues 2–70; AT—A. tumefaciens DnaJ [AAR84666.1] residues 2–70; TC—T. cruzi Tcj3 [AAC18896] residues 2–71; P1—P. falciparum Pfj1 [NP_702750.1] residues 57–125; P4—P. falciparum Pfj4 [BAB17689] residues 2–71; HS—Homo sapiens HSJ1 [NP_001034639] residues 2–69; E1—Mus musculus ERj1 [NP_031895] residues 57–125; E3—H. sapiens ERj3 [NP_057390] residues 21–90; E5—H. sapiens ERj5 [NP_061854] residues 31–99; Con—consensus sequence of the aligned J-domain residues. Con#—consensus numbering used for all sequences in this paper, equivalent to the corresponding E. coli DnaJ residue numbering. *—Proteins of known structure. The numbers on the left-hand side of the alignment represent the positions of the first amino acid in each sequence. Sequences were aligned using ClustalW (Thompson, Higgins, & Gibson, 1994) and then adjusted by hand using available structural data. (B) The ClustalW alignment is represented as an unrooted radial tree using TreeView (Page, 1996). The protein names are indicated at the ends of the branches, and apart from E. coli DnaJ (EcDnaJ), the protein names are indicated by the standard abbreviations. (C) Locations of amino acid residues targeted for substitution shown on a ribbon representation of the known tertiary structure of E. coli DnaJ J-domain (PDB:1XBL). The structure is shown from two different orientations. Helices I–IV are indicated and the highlighted residues are shown as sticks and labelled using the single letter code. The ribbon representations of the structures were rendered using PyMol version 0.98 (DeLano, 2005).