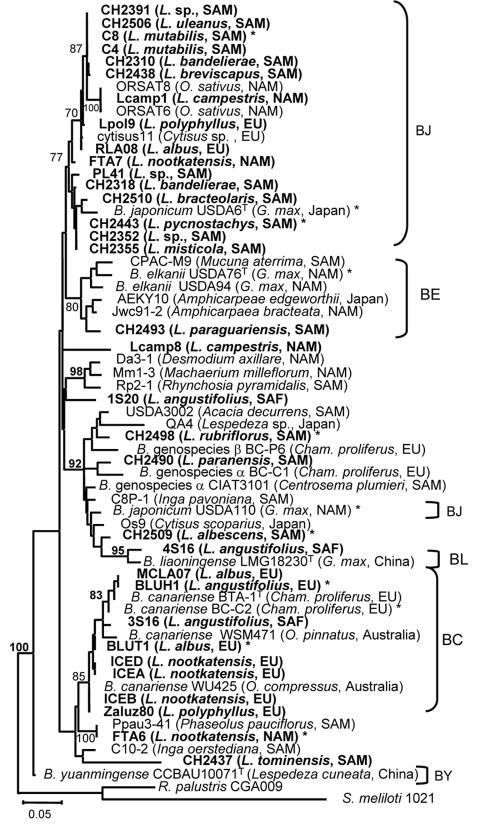
FIG. 3.
ML trees based on partial atpD sequences. The tree shown was constructed unrooted. Lupine strains are indicated in boldface. Host plants and geographical origins of the strains are in parentheses. The scale bar indicates the number of substitutions per site. Bootstrap values indicated are percentages of 1,000 replicates (calculated under distance criteria); they are in boldface when a bootstrap value >70% was found under ML criteria (100 replicates). Abbreviations: BC, B. canariense; BJ, B. japonicum; BL, B. liaoningense; BY, B. yuanmingense; EU, Europe; NAM, North America; SAM, South America; SAF, South Africa. Asterisks indicate strains used for S-H tests (strains for which all markers were sequenced). Reference strains and their accession numbers were as follows: USDA 110, NP_767080; 1S20, AJ891286; BTA-1, AY386739; WSM471, AJ891292; 3S16, AJ891287; WU425, AJ891293; BC-C2, AY386736; BC-P6, AY653751; LMG18230T, AY386752; 4S16, AJ891288; BC-C1, AY386735; CIAT3101, AY653762; Rhodopseudomonas palustris CGA009, NC_005296).