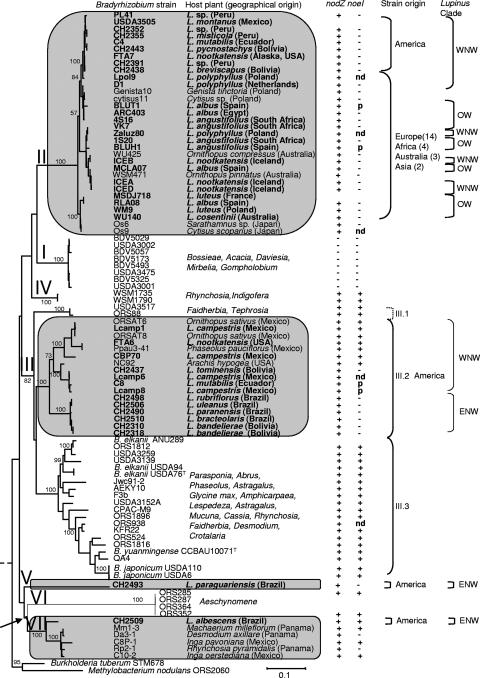
FIG. 4.
ML phylogenetic tree of Bradyrhizobium nodA genes. The tree was constructed by a ML approach described in Materials and Methods. Lupinus strains are indicated in boldface; clades to which they belong are in shaded blocks. The scale bar indicates the number of substitutions per site. Bootstrap values >70% (percentage of 1,000 replicates calculated under distance criteria) are given at the branching nodes. Clades I to IV correspond to the nodA phylogenies described elsewhere (28, 42). Host plant species are indicated on the right side of the tree together with geographical origins in nodA clades containing Lupinus strains. Succeeding columns show results of nodZ and noeI PCR amplifications on test strains, geographical origin of lupine strains, nodA subclades defined by bootstrap values >80%, and finally Lupinus clades corresponding to the phylogeny presented in Fig. 1 including WNW, ENW, and OW. Accession numbers of strains included in this tree are as follows: Bradyrhizobium japonicum USDA 110, NC_004463; B. elkanii USDA 94, U04609; Bradyrhizobium sp. strain ARC403, AJ430731; BDV5029, AJ890291; BDV5057, AJ890311; BDV5173, AJ891168; BDV5325, AJ890290; BDV5493, AJ890312; CBP70, AJ430730; D1, AJ430727; Genista10, AJ430726; NC92, U33192; ORS88, AJ430716; ORS285, AF284858; ORS287, AJ437607; ORS352, AJ438775; ORS364, AJ437613; ORS524, AJ430717; ORS938, AJ430715; ORS1812, AJ430723; ORS1816, AJ430722; ORS1896, AJ430718; 1S20, AJ890296; 4S16, AJ890289; USDA 3001, AJ430713; USDA 3139, AJ430712; USDA 3152A, AJ430711; USDA 3475, AJ430710; USDA 3505, AJ430709; USDA 3517, AJ430708; WSM471, AJ890307; WSM1735, AJ890293; WSM1790, AJ890286; WU140, AJ890313; WU425, AJ890300; VK7, AJ890295; Methylobacterium nodulans ORS2060, AF266748; Burkholderia phymatum STM678, AJ302321. Abbreviations: +, amplified by PCR; −, no PCR product; nd, not determined; p, pseudogene.