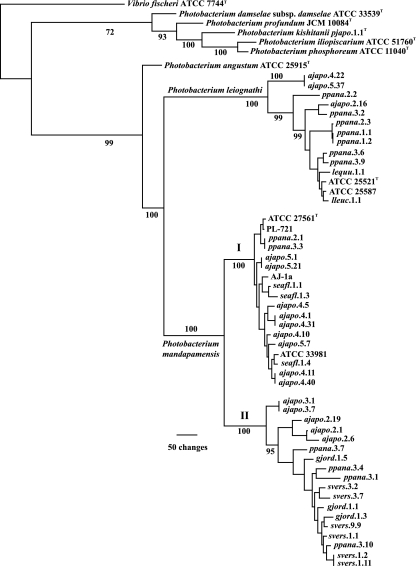
FIG. 1.
Phylogram representing 1 of 56 equally most parsimonious hypotheses resulting from combined multilocus analysis of gyrB and luxAB(F)E gene sequences of Photobacterium species (1,393 informative characters; length, 3,491; consistency index [CI], 0.578; retention index [RI], 0.879). A strict consensus of the 56 trees reduces resolution only among smaller clades near the tips of the tree. The separation of P. leiognathi and P. mandapamensis results primarily from differences in lux operon sequences, as described previously (1), and was confirmed here through analysis of ribBHA sequences (Fig. 3). Numbers at the nodes indicate Jackknife resampling values; some values were omitted for clarity. Representative strains from different specimens of S. versicolor were used here. Strains whose gyrB and luxAB(F)E sequences were identical and that were therefore excluded from this analysis are gjord.1.4, gjord.1.7, and gjord.1.9 (identical to gjord.1.1); gjord.1.8 (identical to gjord.1.3); gjord.1.10 (identical to gjord.1.5); and ppana.3.14 (identical to ppana.3.1 [see the text]). Strains with identical rep-PCR profiles and therefore not sequenced or, if sequenced, not used in this analysis due to sequence identity are gjord.1.2 (identical to gjord.1.1), gjord.1.6 (identical to gjord.1.4), ppana.1.3 through ppana.1.20 (identical to ppana.1.1 and ppana.1.2), ppana.2.5 through ppana.2.12 (identical to ppana.2.3), and ppana.3.5 (identical to ppana.3.3). For strains from specimen Ajapo.2, representatives of each strain type, ajapo.2.1, ajapo.2.6, ajapo.2.16, and ajapo.2.19, were sequenced and used in this analysis. Roman numerals I and II refer to P. mandapamensis clade I and clade II, respectively (see the text).