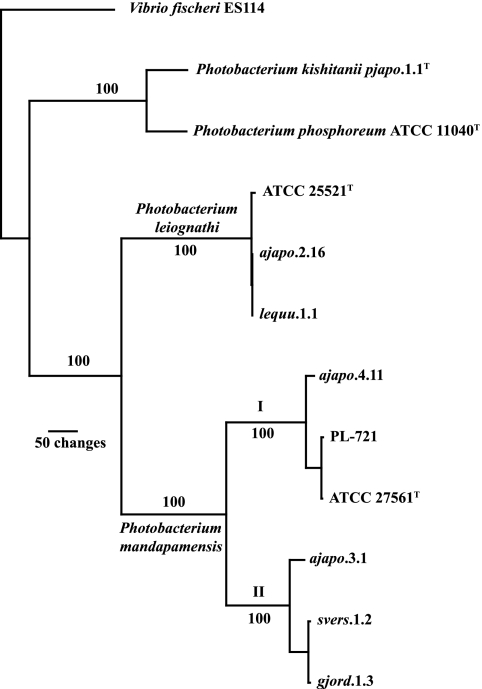
FIG. 3.
Phylogram of the single most parsimonious tree resulting from analysis of the genes ribB, ribH, and ribA (868 informative characters; length, 1,603; CI, 0.812; RI, 0.861). Sequences were aligned by inferred amino acid sequences and analyzed using the branch-and-bound algorithm as implemented in PAUP* (39). Jackknife resampling values appear at the nodes and were calculated using 1,000 branch-and-bound replicates with 34% deletion, emulating Jac resampling. Roman numerals I and II refer to P. mandapamensis clade I and clade II, respectively. Compare the figure with Fig. 1.