Abstract
Recent advances in molecular genetics impact the health care and outcome of patients with acute lymphoblastic leukemia (ALL). BCR-ABL, a common molecular defect in adult ALL, is a valuable tumor marker whose detection influences prognosis and clinical management decisions. Molecular methods such as fluorescence in situ hybridization (FISH), reverse-transcriptase polymerase chain reaction (rtPCR), and real-time quantitative rtPCR can be used to detect the chimeric BCR-ABL gene or its transcripts. These molecular assays improve our ability to measure residual disease and to estimate risk of relapse. On the horizon are gene expression profiles that will likely provide additional information beyond what is obtainable with current clinical and laboratory approaches.
In 1960, Nowell and Hungerford 1 reported the discovery of what came to be known as the Philadelphia chromosome (Ph1) in association with chronic myelogenous leukemia (CML). Their short report entitled “Minute chromosome in human chronic granulocytic leukemias” was the first to demonstrate a leukemia-specific genetic abnormality. In the ensuing decades, the pathology of the Ph1 has been studied and it is now known to represent an abnormally shortened (derivative) chromosome 22 resulting from translocation with chromosome 9. 2 The t(9;22) is found in over 90% of CMLs, in a lesser proportion of acute lymphoblastic leukemias (ALL) or biphenotypic acute leukemias, and in rare cases of de novo acute myelogenous leukemia (AML).
The break on chromosome 22q11.2 usually occurs in the major breakpoint cluster region (M-BCR), in the minor breakpoint cluster region (m-BCR), or rarely at other nearby sites. The break on chromosome 9q34 involves the ABL gene, named after Abelson murine leukemia virus where a viral version of the ABL gene was first discovered. The translocation brings the 5′ end of the BCR gene into juxtaposition with the tyrosine kinase domain of the ABL gene to produce a hybrid gene retaining tyrosine kinase activity. 3 Depending on whether the M-BCR or m-bcr breakpoint is involved in the translocation, transcription of the hybrid gene results in chimeric mRNA encoding a 210 kd BCR-ABL fusion protein or a 190 kd BCR-ABL fusion protein, respectively. The reciprocal ABL-BCR translocation also forms a chimeric gene that is capable of being transcribed, but the pathological significance of this reciprocal chimeric gene product is uncertain.
Laboratory Tests for t(9;22)
Conventional cytogenetics is the recommended test for detecting t(9;22) in newly diagnosed leukemia patients. Chromosome banding analysis has the advantage of high specificity and an ability to detect alternate or additional cytogenetic defects that are valuable in diagnosis and prognosis. However, cytogenetic analysis requires viable marrow cells or more than 10% blasts in the peripheral blood to reliably culture the cells and visualize metaphases. Occasionally fibrosis interferes with marrow aspiration, yielding few analyzable metaphase cells. The number of cells examined determines the sensitivity of karyotyping. A typical examination of 20 cells carries a sensitivity of one in 20, or 5%. Cryptic t(9;22) occurs in about 5% of CML cases and also in a small proportion of ALLs, resulting in false negative karyotypic interpretation in metaphase spreads of cells that actually contain the translocation at the molecular level. 4
Fluorescence in Situ Hybridization
Fluorescence in situ hybridization (FISH) allows detection of the BCR-ABL translocation in either metaphase or interphase cells. Because interphase cells are suitable, FISH can be applied directly to blood leukocytes and other non-dividing cells. Moreover, FISH detects cryptic and complex BCR-ABL rearrangements such as three-way translocations or breaks outside of the usual major and minor cluster regions. 4 Dual color FISH utilizes two probes, one of which hybridizes to the 5′ end of the BCR region and the other to the 3′ end of the ABL region. Fluorescent microscopy is used to visualize these probes which are typically labeled with red and green fluorochromes to produce two red and two green spots representing the two copies of chromosomes 9 and 22 in normal cells. In cells harboring a BCR-ABL translocation, a red and green probe are juxtaposed to produce a yellow fluorescent signal. Typically, 200 interphase or metaphase nuclei are evaluated, yielding a sensitivity of about 1 in 200, or 0.5%.
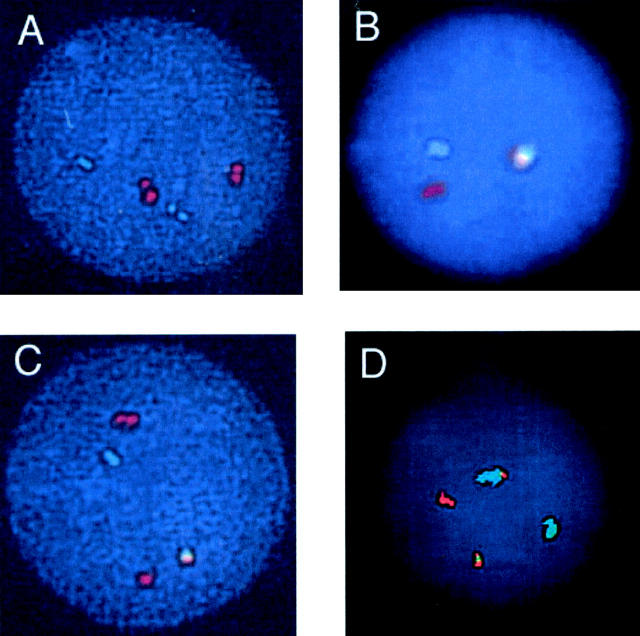
This classic dual-color single-fusion FISH (S-FISH) assay is highly accurate for analyzing metaphases, but is hampered in its application to interphase cells by the coincidental overlap of BCR and ABL signals in about 4% of normal nuclei. 5 Because it is prone to false positivity, quantification below 10% is generally considered unreliable. 6, 7, 8, 9 To improve assay specificity, alternate strategies were applied in the design of newer DNA probes. For example, extra-signal FISH (ES-FISH) employs a larger 650-kb ASS-ABL probe (Vysis, Downer’s Grove, IL) which targets an area spanning the ABL gene and the adjacent arginino-succinate synthetase (ASS) gene. Since this probe spans both sides of the ABL breakpoint cluster regions, assay specificity is improved. In a validation study conducted at our institution, 30 non-leukemic patients were assayed for the presence of the BCR-ABL fusion using interphase ES-FISH. The background level of fusion signals in marrow samples was less than or equal to 5% with a confidence limit of 95%, indicating a marginal improvement in assay specificity. ES-FISH is also touted for its ability to distinguish M-BCR from m-BCR based on the appearance of the spot pattern. 10 (See Figure 1 .)
Figure 1.
FISH using the Vysis Extra Signal (ES) probe reveals a normal result yielding two green chromosome 22 and two red chromosome 9 signals (A); one green, one red, and one red-green fusion signal, representing a pattern that could be artifact due to coincidental juxtaposition of red and green signals, or it could represent BCR-ABL translocation with concomitant loss of chromosome 9 material proximal to the breakpoint, or finally it could represent an insertion of bcr into the chromosome 9 long arm at the abl locus (B); typical findings in CML (p210 breakpoint) yielding one green, one red, one residual red, and one red-green fusion signal (C); and typical findings in ALL (p190 breakpoint) yielding one green, one red, and two red-green fusions (D).
Another strategy to improve assay specificity is dual-fusion FISH (D-FISH). This involves applying probes that span the common breakpoint regions of both ABL and BCR genes so that one may visualize both the BCR-ABL and the ABL-BCR fusion signals. In a commercial version of this system (Vysis, Downers Grove, IL) the large ASS-ABL probe mentioned above is combined with a large probe spanning a distance of about 1.5 mb on both sides of the BCR gene, thus providing a second confirmation of the translocation and reducing the rate of false positives to less than 0.5%. 4, 11, 12, 13 In addition, D-FISH, like S-FISH and ES-FISH, can detect cryptic and complex BCR-ABL translocations that are missed or undecipherable by conventional cytogenetics. 4
Southern Blot Analysis
Southern blot analysis reliably identifies BCR gene rearrangement using probes targeting either the M-bcr or m-bcr breakpoint. While this is quite helpful in confirming the BCR defect associated with CML or ALL, the Southern blot method suffers from high cost and slow turnaround time. In addition, the assay is not sensitive for detecting minimal residual disease since tumor levels below about 5% are not detectable.
Amplification Technology
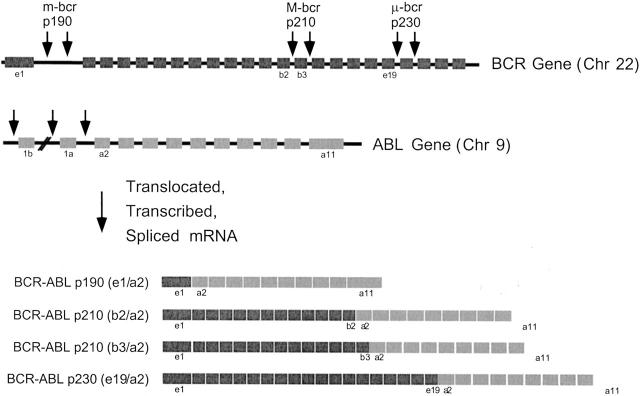
Reverse transcriptase polymerase chain reaction (rtPCR) is the most sensitive method described to date for detecting BCR-ABL. Instead of targeting chromosomal DNA, the assay targets the more abundant chimeric RNA transcripts produced from the fused genes. Chromosomal DNA is an impractical target not only because it is less abundant than RNA but also because the breakpoint regions span such large segments of intronic DNA that multiple PCR primer sets would be required to detect every possible translocation. On the other hand, the chimeric RNA is remarkably homogeneous from case to case, thus allowing reliable detection of nearly all disease-associated translocations (Figure 2) .
Figure 2.
Diagrammatic representation of the BCR and ABL genes and the corresponding BCR-ABL fusion transcripts. Arrows designate three different breakpoint cluster regions in the BCR gene. Breaks in the M-BCR are most commonly associated with CML, the m-BCR with ALL, and the μ-BCR with chronic neutrophilic leukemia. ABL breakpoints usually occur between exons 1a and 2, resulting in fusion transcripts containing the tyrosine kinase domain of the ABL gene and variable portions of the BCR gene.
Amplification assays are capable of detecting one affected cell among 100,000 or so normal cells. The exceptional sensitivity of rtPCR makes it well suited for assessing minimal residual disease following therapy. A downside that has hampered its implementation in clinical laboratories is the possibility of false positive results due to contamination of a negative specimen by amplicons produced from prior positive amplification reactions. 14 Recent technological improvements such as chemical destruction of wayward amplicons (eg, uracil-N-glycosylase system), or real-time detection of products to avoid amplicon manipulation, help minimize the possibility of amplicon contamination.
Another cause for concern is physiological positive results that mimic tumor-associated positive results. Spurious positive results for BCR-ABL rtPCR are identified in blood samples from up to 75% of normal individuals. 15, 16 The majority of these normal individuals express only minute amounts of BCR-ABL transcripts that were detected by highly sensitive assays capable of detecting 1 in 100 million tumor cells. Typical clinical assays are a thousand-fold less sensitive and very rarely yield positive results in healthy individuals. Nevertheless, it is advisable to repeat any positive test when looking for residual disease and to compare the size of the amplicons with those known to be produced from the patient’s tumor. Newly available quantitative rtPCR assays now make it possible to assess trends in the BCR-ABL load over time. These quantitative tests are further described in the section on minimal residual disease.
False negative amplification results are also a concern, particularly since RNA is the substrate for rtPCR assays, and RNA is notoriously subject to degradation by ubiquitous RNase enzymes. It is therefore advisable to stabilize RNA immediately on sample collection and to use appropriate control tests to confirm that amplifiable cDNA is present in each patient specimen.
Gene Expression Profiles and Microarray Analysis
Microarrays have been used to survey gene expression in ALL samples. A major advantage of array technology is the ability to evaluate expression of thousands of genes simultaneously. In a typical analysis, RNA is extracted from the patient sample and converted to labeled cRNA or cDNA before being applied to an array of complementary probes. Yeoh et al 17 reported using 12,600 probe sets to study 327 diagnostic marrow samples from childhood ALL patients. They could distinguish BCR-ABL cases from other forms of ALL, and they also identified additional subsets of patients at high risk of relapse based on their gene expression profiles. The findings are promising and will be followed by more studies aimed at defining a smaller panel of probe tests that is informative for diagnosis, classification, prognosis, and predicting response to therapy.
Prevalence of BCR-ABL in Acute Leukemia
The reported prevalence of the Ph1 in adult ALL is 20 to 23% by conventional cytogenetic testing. 18, 19, 20 More sensitive rtPCR techniques reveal a prevalence of 27 to 30%. 20, 21, 22 In a study by Gleissner et al, 23 rtPCR testing for BCR-ABL failed or produced false negatives in 19 of 212 patients (9%). This was due to insufficient material in 10 cases, ambiguous results in Ph1-negative patients in six cases, and false negative test results in three cases. Cytogenetic analysis failed or was falsely negative in 48 patients (23%) including 32 non-analyzable and 16 false negative test results. Despite the possibility of failure, chromosome banding analysis remains the best first-line genetic test to assess any new acute leukemia because it screens for t(9;22) as well as for alternate or additional genetic defects. When negative or inconclusive results are obtained by cytogenetics, then FISH or rtPCR are recommended to determine whether BCR-ABL is present.
In childhood ALLs, the incidence of BCR-ABL translocation is much lower at only 2 to 4% depending on the method used for identification. 24, 25, 26, 27 BCR-ABL transcripts are also present in 31 to 35% of biphenotypic acute leukemia in adults and children, 22, 28 in 6% of de novo adult AML, 22 and in 1% of childhood AML. 29
Clinical and Immunological Correlates at Initial Diagnosis
The majority of BCR-ABL positive adult and childhood ALLs have a typical pre-B cell immunophenotype with co-expression of CD10 (CALLA). Indeed, when results from five different studies were combined, including 170 adults and children, 94% of BCR-ABL positive ALL had a pre-B cell immunophenotype. 20, 24, 26, 30, 31 In a study by Gleissner et al 23 on 875 adult ALL patients, the prevalence of BCR-ABL positivity among CD10-expressing adult pre-B ALL was 37% whereas in the CD10-negative cohort it was only 2%. The incidence of BCR-ABL in T-cell leukemias varies among studies from 0% to 5%. 20, 23, 24, 26, 30, 31
Coexpression of myeloid markers, as defined by CD13 or CD33 in at least 20% of blasts, is found in 27 to 29% of BCR-ABL positive adult and childhood ALL. 23, 30, 32 Myeloid markers are more frequently seen in BCR-ABL positive than in BCR-ABL negative ALL. 23, 26 There was no correlation between myeloid co-expression and p190 versus p210 breakpoint, 20, 30 nor was there a correlation with WBC count, blast count, hemoglobin, or hematocrit. 31 Some investigators suggest that myeloid antigen expression has an adverse impact on survival in BCR-ABL positive ALL patients while others do not. 19, 30, 32
Studies comparing the presenting clinical features of BCR-ABL positive versus negative ALL have also produced conflicting results. Some investigators found no significant difference in age, presenting WBC count, percentage of blasts, splenomegaly, or hepatomegaly 20, 26, 30 while others found that BCR-ABL positive children 24, 25 and adults 23 had higher WBC counts and older age. Recently, Gleissner et al reported that hepatosplenomegaly was more common in BCR-ABL negative adult ALL patients. 23
The Significance of p210 versus p190 Breakpoint
An advantage of rtPCR and of certain FISH assays is the ability to differentiate p210 from p190 forms of BCR-ABL. Nearly all CML patients have a p210 breakpoint, and they retain this genotype if their disease progresses to blast crisis. In contrast, the majority of de novo Ph1-positive acute leukemias harbor a p190 breakpoint. Therefore, patients whose acute leukemia harbors a p210 breakpoint should be evaluated to distinguish de novo acute leukemia from blast crisis of CML. This distinction can be difficult or impossible unless the patient had a prior white cell count or signs and symptoms of CML, or unless the patient reverts to chronic phase after treatment.
Among children and adults with Ph1 positive ALL, approximately 75% have p190 and 25% have a p210 breakpoint in the BCR locus. 20, 23, 26, 30 Approximately 3% of adult ALL patients express both p210 and p190 fusion transcripts. 23 This is explained by alternative splicing whereby M-BCR breaks produce both p210 and, at a lower level, p190 transcripts. 23, 33 Most studies show no correlation between the type of breakpoint and clinical parameters or prognosis, 20, 26, 30, 34 while a few studies showed patients with p210 transcripts were likely to be older. 23, 35 Some investigators found a tendency for p210 positive patients to do better 36, 37, 38 or worse 23 than p190 patients. In Ph1-positive AML patients, the p210 breakpoint is present in about half while the rest have a p190 breakpoint. AMLs harboring a p190 breakpoint usually have a monocytic (FAB M4 or M5a) phenotype. 18, 39
As noted above, alternative splicing can result in p190 transcript production from template DNA having a p210 breakpoint, and therefore CML or ALL (M-BCR breakpoint) patients may have low levels of p190 transcripts, usually not comprising over 10% of the total BCR-ABL mRNA. 33, 40, 41, 42, 43 In one study, p210 transcripts were detected in all CML patients in accelerated phase or blast crisis, and coexisting p190 transcripts were found in 8%. 42 None of these patients had p190 mRNA at the time of initial diagnosis. The association of p190 detection with relapse was reported by Serrano et al in a study of 55 CML patients who underwent marrow transplant. 43 All 14 patients who relapsed had become p190 positive by the time of cytogenetic relapse. In contrast, the 41 patients who remained in remission consistently tested negative for p190 but not necessarily for p210. Radich et al 37 studied 36 ALL patients at multiple timepoints after marrow transplant. Among 23 patients with at least one positive rtPCR result, 8 had p210 alone, 10 had p190 alone, and 5 had both types of BCR-ABL transcripts. The patients with p190 transcripts detectable after transplant were more likely to relapse than were patients with p210 alone or with undetectable BCR-ABL. This could be because p190 protein has stronger tyrosine kinase activity and is associated with a more aggressive leukemia in animal model systems.
The Impact of BCR-ABL on Prognosis of ALL
While early reports showed a lower response to induction chemotherapy in Ph1-positive patients, 24 this has not been the case with the use of modern therapies. In a large study of 1322 children with ALL, nearly all Ph1-positive (97%) and Ph1-negative (98%) patients achieved complete remission after induction therapy. 25 But those who had Ph1-positive leukemias were more likely to relapse. The estimated 4-year event-free survival was only 20% for Ph1-positive as compared with 76% for Ph1-negative patients. The four-year overall survival was 56% for Ph1-positive and 85% for Ph1-negative patients. The poor prognosis in children with Ph1 positive ALL was confirmed by Schrappe et al, 30 where complete remission was achieved in 90% but the 4-year event-free survival was only 38%, and 4-year overall survival was 48%.
Analogous to the situation in childhood ALL, young adults consistently have a worse prognosis when their ALL harbors BCR-ABL translocation. Gleissner et al 23 found that 68% of BCR-ABL positive adult ALL patients achieved complete remission compared to 85% of BCR-ABL negative patients, with a higher frequency of early relapses in BCR-ABL positive patients. The median overall survival was 11 months for BCR-ABL-positive and 30 months for BCR-ABL-negative patients. The 3-year overall survival was 15% for BCR-ABL-positive and 47% for BCR-ABL-negative patients. Kantarjian et al, using a hyper-CVAD regimen to treat adult ALL, confirmed that the complete remission rate for Ph1-positive patients was not different from that of the whole cohort (91%), but their 5-year survival was only 7% as compared to 39% for the entire study population. 19 The findings may not extend to the elderly, however, based on work from Onciu et al 44 showing that ALL patients over 59 years of age did not display an association between Ph1 and poor prognosis.
The prognosis of biphenotypic acute leukemia (BAL) is worse than either ALL or AML. In a study of 23 adult BAL patients by Legrand et al, complete remission was achieved by 48% as compared to 65% of adult AML and 81% of adult ALL patients. 22 The median overall survival was 7.5 months for BAL, 11 months for AML, and 12 months for ALL. Killick et al, 45 in a report of 25 adult and pediatric BAL patients, found that prognosis was strongly related to t(9;22) status and age less than 15 years. The two-year overall survival for adults was 17% and for children was 75%. There was no difference in survival between children with Ph1-negative BAL and those with Ph1-negative AML or ALL suggesting that the worse prognosis in BAL may be related to BCR-ABL positivity.
Impact of BCR-ABL on Selecting Therapy for ALL
Because of the high rate of relapse and low overall survival in BCR-ABL positive acute leukemia patients, detection of this genetic abnormality is considered an indication for allogeneic BMT in first remission. 27, 46, 47 In a report of 30 children with Ph1-positive ALL, 29 achieved complete remission. 25 Fifteen of these patients underwent allogeneic BMT (five were related, seven were unrelated, and three were unspecified); 10 of these transplants occurred at the time of first complete remission. After follow-up for a median of 39 months, there were 6 event-free survivors among these 10 patients and only 2 event-free survivors among the other 20 patients, underscoring the utility of BMT in first remission.
In another study involving 61 children with BCR-ABL positive ALL, 28 underwent marrow transplant (23 were related, 5 were unrelated). 30 The four-year event-free survival for the whole group was 38%, 61% for patients who were transplanted, and 28% for patients who received only chemotherapy. Unrelated donor transplantation was associated with lethal toxicity in four of five patients. On the other hand, 15 of the 19 patients who received matched-related marrow transplant remained in first complete remission, and their four-year event-free survival was 83%.
As further support for transplant of Ph1-positive ALL during first remission, a study of 32 such children was conducted. 26 Thirty achieved complete remission, of whom eight then received allogeneic marrow, three received autologous marrow, and 19 received chemotherapy alone. Subsequently, three of the latter patients received allogeneic marrow and one received autologous marrow. All patients relapsed except six of the eight patients who received allogeneic marrow during first remission. There were no long-term survivors among relapsed patients despite treatments including allogeneic transplant.
Two long-term prospective studies confirm that adult ALL has a poor prognosis. 48, 49 The highest risk was in patients who were over 35 years of age, Ph1-positive, had leukocyte counts above 30 × 10(9)/L, had null ALL or undifferentiated leukemia, or required longer than four weeks to achieve complete remission. A few studies specifically addressed the role of allogeneic BMT in Ph1-positive patients. Improved survival data were reported by Forman et al 50 in 10 such patients of whom six were alive and well for a median of 19 months. Snyder et al 51 showed that 23 Ph1-positive adult ALL patients who were transplanted in first remission had improved disease-free survival (65% at 3 years). Mitterbauer et al 21 reported that allogeneic transplant was more effective than chemotherapy alone in reducing BCR-ABL levels and in achieving long term remission. Among adults treated with stem cell transplant in first remission for Ph1 ALL, Dombret et al 38 showed that those who achieved negativity for BCR-ABL before transplant had improved remission duration and survival after transplant.
Gleevec Therapy for BCR-ABL Leukemias
Imatinib (Gleevec), which targets the ABL tyrosine kinase hyperactivity of the BCR-ABL oncoprotein, has been shown to be effective in the treatment of CML or CML in myeloid or lymphoid blast crisis. 52, 53 Unfortunately, Gleevec resistance sometimes develops after an initial response. A study by Gorre et al 54 showed that Gleevec resistance in nine CML patients was associated with reactivation of tyrosine kinase activity. In three patients, there was BCR-ABL chimeric gene amplification, while the other six patients acquired a point mutation in the tyrosine kinase domain of BCR-ABL (Thr315Ile).
Gleevec is also temporarily effective in treatment of BCR-ABL-positive ALL as shown in a recent report of 21 patients who had initially relapsed or failed to respond to standard chemotherapy. 55 With Gleevec therapy, 13 of these patients were classified as good responders (12 achieved complete hematological remission and one achieved partial remission with peripheral hematological recovery). Marrow aspirates of 9 of the 21 patients (5 good responders and 4 resistant patients) were obtained before and after therapy. Direct sequencing of cDNA representing a 714 bp segment of ABL encoding the ATP-binding site and the kinase activation loop showed an acquired point mutation at position 1127 (Glu255Lys) in 6 patients (4 good responders and 2 resistant patients). A seventh resistant patient acquired a point mutation at nucleotide 1308 (Thr315Ile). Other reports have confirmed the acquisition of point mutations in the ATP-binding domain of BCR-ABL in ALL and CML patients who became resistant to Gleevec. 56 Screening for these mutations may allow therapy to be altered before frank relapse.
Minimal Residual Disease Detection
Detection of minimal residual disease is beneficial to patients with ALL, and advances in laboratory technology provide new and more powerful ways to detect low level disease. 57, 58, 59 Most investigators have applied one of three approaches: 1) PCR amplification of rearranged IgH or TCR genes using custom-designed tumor-specific primers and probe; 2) multiparameter flow cytometry; or 3) Quantitative PCR of tumor-associated translocation breakpoints. All three approaches have merit. Targeting the IgH or TCR genes is feasible in nearly all cases of ALL, but the development of customized IgH or TCR probes is costly and labor-intensive. Flow cytometry is quite helpful assuming that the leukemic clone has an aberrant phenotype not prevalent in normal cells in the sample.
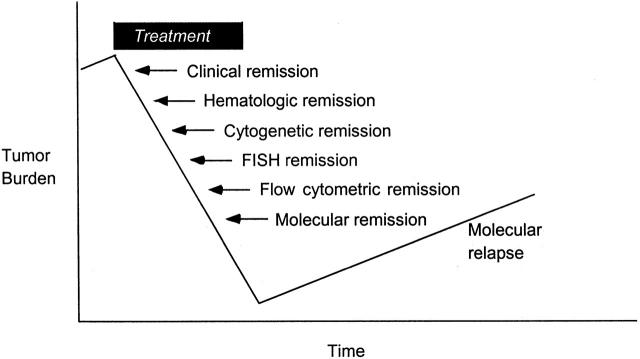
Analysis of tumor-specific translocations is a promising alternative for evaluating minimal residual disease in those patients whose tumor harbors a translocation for which a reliable laboratory assay is available. 60, 61 BCR-ABL is an appealing target considering that a significant proportion of ALLs harbor the translocation (about 28% of adult ALL; 3% of childhood ALL), and sensitive assays are available for detecting it by rtPCR. Such amplification assays are the most sensitive of all laboratory approaches, followed by flow cytometry, FISH, and cytogenetics, with associated detection thresholds of about 1 in 1 million, one in several thousand, one in several hundred, and 1 in 20, respectively. (See Figure 3 .)
Figure 3.
Laboratory tests are capable of detecting tumor burden at different levels. Up to a billion leukemic cells may remain in a patient who is in hematological remission, underscoring the importance of using more sensitive assays to detect and measure minimal residual disease. Theoretically, rising levels permit early intervention so that the number of cell divisions is minimized and the accompanying risk of secondary genetic events is likewise reduced.
Real-time quantitative rtPCR techniques are now available that permit precise measurement of chimeric transcripts. Levels can be reported either in absolute terms or relative to a housekeeping transcript. Recent studies using this technology to detect BCR-ABL transcripts have shown good correlation with the results of cytogenetic analysis, FISH, Southern blot analysis, and conventional nested rtPCR. 62, 63, 64 Various platforms such as the ABI Prism 7700 (Applied Biosystems, Foster City, CA) or the LightCycler (Roche Diagnostics, Indianapolis IN) were able to reproducibly detect one positive cell from among 10,000 to 100,000 normal cells. 62, 63, 64, 65, 66, 67, 68, 69, 70
Investigators have consistently demonstrated the prognostic value of molecular analysis of BCR-ABL transcripts in the management of ALL patients. After successful marrow transplantation, ALL patients tend to become rtPCR negative within six weeks. 71 Persistence of BCR-ABL transcripts is associated with a high risk of relapse, while those who remain rtPCR negative have a good prognosis. 37, 71, 72, 73 Disease-free survival is related to the rtPCR status after marrow transplantation regardless of the presence of residual disease before transplantation. 73, 74
The predictive value of molecular follow-up has been confirmed in a study by Radich et al 37 of 36 adults and children who were transplanted for Ph1-positive ALL. Patients in whom rtPCR was positive on one or more occasions after transplant were more likely to relapse, and the unadjusted relative risk of relapse for a positive test was 5.7. A positive p190 result was particularly indicative of a high risk of relapse while p210 positivity was not. Indeed, of 10 transplant patients who were rtPCR positive and relapsed, 9 had p190 either alone (7 patients) or in concert with p210 (2 patients). In a more recent study by Radich et al, 72 the ABI Prism 7700 real-time rtPCR platform was used to quantitate BCR-ABL in posttransplant samples from 12 CML patients who relapsed and 73 patients who remained in remission. BCR-ABL levels were a significant prognostic indicator in these divergent outcome groups.
There is reasonably good correlation between BCR-ABL rtPCR results from blood and marrow samples collected at the same time. Lin et al reported complete concordance in blood and marrow samples obtained on 23 occasions from CML patients following allogeneic BMT or interferon therapy. 75 Positive cases showed good correlation in the number of BCR-ABL transcripts per microgram of RNA (r = 0.99). Radich et al found a 91% correspondence in BCR-ABL rtPCR results between blood and marrow samples obtained on 605 occasions from CML patients who had undergone allogeneic BMT. 72 Evaluation of the occasional discordances showed marrow alone was positive in 36 cases while blood alone was positive in 18 cases. In an analogous study of ALL patients, Radich et al 37 reported a 74% concordance in samples obtained on 31 occasions from ALL patients who had undergone allogeneic BMT, with the few discrepancies involving marrow-only positivity in five cases and blood-only positivity in three cases. The findings suggest that either blood or marrow samples may be suitable for follow up of BCR-ABL associated leukemias. Surprisingly, archival formalin-fixed paraffin or acrylate-embedded marrow biopsies have recently been shown to be suitable sample types for BCR-ABL rtPCR. 76
It is theorized that minimal residual disease after marrow transplantation could be controlled with relatively modest intervention. A recent report showed that residual BCR-ABL transcripts in two ALL patients were rendered undetectable following acute graft versus host disease. Either donor lymphocyte infusion (DLI) or rapid reduction of immunosuppression caused this graft versus host effect, and it was associated with lasting disease remission. 77 Similarly, it has been shown that CML patients who achieve complete remission after DLI have prolonged survival. In a study of 39 CML patients who achieved cytogenetic remission with DLI, the overall probability of survival was 87% at 1 year, 76% at 2 years, and 73% at 3 years (median follow-up of 40 months after DLI). 78 The use of rtPCR positivity as the criterion for relapse following allogeneic BMT and as the indication for DLI has been investigated. Mughal et al presented data on 20 CML patients in molecular relapse who were treated with DLI. 79 After a median follow-up of 42 months, these patients were in continuous molecular remission. Their actuarial probability of survival at 10 years after BMT was 100% and their probability of relapse was 0%. The actuarial probability of survival at 10 years for 63 control CML patients who had been continuously PCR negative after allogeneic BMT was 97% (median follow-up of 8.4 years after allogeneic BMT). The availability of such effective measures for detecting and dealing with minimal residual disease is promising.
Currently there are no definitive guidelines on when or how frequently to monitor BCR-ABL for purposes of minimal residual disease detection in ALL patients. If we extrapolate from the extensive literature on monitoring BCR-ABL in CML patients, we infer that the type of test and frequency of its application depends on which therapy is used and on its curative versus palliative intention. 73 CML treated with hydroxyurea, busulfan, or other conventional non-curative therapy relies on cytogenetics to monitor residual disease and to detect markers of progression, while more sensitive molecular tests such as FISH are only marginally more informative, and rtPCR is probably not needed at all. 9 When interferon therapy is used to treat CML, the European consortium recommends that once the number of Ph1-positive cells has fallen below 10% by karyotype then quantitative rtPCR should be performed every 3 months. 81 Interferon should be continued until the BCR-ABL/ABL ratio is lower than 0.02%. 82 After stem cell transplant, the European consortium recommends quantitative rtPCR be done every 4 weeks for as long as BCR-ABL is detectable, then at intervals of 3 months and, after 1 year of undetectable levels, at up to 6-month intervals. 81 If levels increase, immediate retesting helps to confirm and direct intervention. This group has defined molecular relapse as a 1 log (10-fold) or greater increase in BCR-ABL determined by a minimum of three consecutive quantitative rtPCR tests regardless of the interval between these tests. Lin et al defined molecular relapse as >50 transcripts per microgram of RNA or increasing values on serial testing. 83 A more recent report from the same institution by Olavarria et al suggested the following criteria: A) the BCR-ABL/ABL ratio is >0.02% on three consecutive occasions over 1 month apart; or B) the ratio is >0.05% on two consecutive samples; or C) the transcript number is rising in three consecutive samples, with at least two samples having ratios above 0.02%. 84
The National Comprehensive Cancer Network (NCCN) recommends less frequent monitoring of residual disease in CML. 85 According to their recent practice guidelines, after hematological remission is achieved following allogeneic marrow transplant, cytogenetics and rtPCR should be done every 6 months for the first 2 to 3 years and yearly thereafter. 85 If karyotype is positive at any timepoint, the patient should be treated for relapse with interferon or donor lymphocyte infusion (DLI). If rtPCR is positive but karyotype is negative, the patient should be observed and retested every 6 months for 2 years and then every 12 months. After treatment of relapse, the guidelines recommend rtPCR testing every 6 months for 2 years and yearly thereafter, with cytogenetic testing at any rtPCR-positive timepoint. Several clinical trials are underway to further determine the utility and recommended frequency of laboratory monitoring of CML, particularly now that Gleevec has been added to the armamentarium. Of note, one mechanism of Gleevec resistance is amplification of the BCR-ABL chimeric gene, leading to overexpression of the fusion transcript rather than the anticipated decrease expected with successful therapy. 54
Conclusion
Patients with pre-B ALL should be tested for BCR-ABL at the time of diagnosis to identify those who have the translocation and who therefore carry a worse prognosis and are candidates for treatment with intensive induction regimens and marrow transplantation. Transplantation during first remission appears to benefit such patients. Studies are underway to assess alternative treatment programs such as Gleevec for their role in disease management.
Karyotype should be initially performed on all suspected ALL patients. While karyotyping has the advantage of being able to detect a large variety of genetic alterations, it is also vulnerable to false negative results. FISH and rtPCR are helpful for detecting occult t(9;22) and for differentiating p190 and p210 breakpoints. Amplification strategies are quite sensitive and thus well suited for follow-up to detect minimal residual disease. One caveat, however, is that trends in BCR-ABL test results rather than a value obtained at a single timepoint should be taken into consideration before making clinical management decisions. The recent advent of quantitative BCR-ABL rtPCR testing permits precise measurement of low levels of BCR-ABL that appear to reflect trends in tumor burden.
Address reprint requests to Margaret L. Gulley, M.D., Department of Pathology, Brinkhous-Bullitt Building, University of North Carolina, Chapel Hill, NC 27599-7525. E-mail: margaret_gulley@med.unc.edu.
References
- 1.Nowell PC, Hungerford DA: A minute chromosome in human chronic granulocytic leukemia. Science 1960, 132:1497 [Google Scholar]
- 2.Rowley JD: A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243:290-293 [DOI] [PubMed] [Google Scholar]
- 3.Davis RL, Konopka JB, Witte ON: Activation of the c-abl oncogene by viral transduction or chromosomal translocation generates altered c-abl proteins with similar in vitro kinase properties. Mol Cell Biol 1985, 5:204-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelz AF, Kröning H, Franke A, Wieacker P, Stumm M: High reliability and sensitivity of the bcr/ABL1 D-FISH test for the detection of BCR/ABL rearrangements. Ann Hematol 2002, 81:147-153 [DOI] [PubMed] [Google Scholar]
- 5.Chase A, Grand F, Zhang J-G, Blackett N, Goldman M: Factors influencing false positive and negative rates of BCR-ABL fluorescence in situ hybridization. Genes Chromosomes Cancer 1997, 18:246-253 [PubMed] [Google Scholar]
- 6.Dewald GW, Schad CR, Christensen ER, Tiede AL, Zinsmeister AR, Spurbeck JL, Thibodeau SN, Jalal SM: The application of fluorescent in situ hybridization to detect Mbcr/abl fusion in variant Ph chromosomes in CML and ALL. Cancer Genet Cytogenet 1993, 71:7-14 [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Isidoro M, Tabernero MD, Garcia JL, Najera ML, Hernandez JM, Wiegant J, Raap A, San Miguel J, Orfao A: Detection of the Mbcr/abl translocation in chronic myeloid leukemia by fluorescence in situ hybridization: comparison with conventional cytogenetics and implications for minimal residual disease detection. Hum Pathol 1997, 28:154-159 [DOI] [PubMed] [Google Scholar]
- 8.Werner M, Ewig M, Nasarek A, Wilkens L, von Wasielewski R, Tchinda J, Nolte M: Value of fluorescence in situ hybridization for detecting the bcr/abl gene fusion in interphase cells of routine bone marrow specimens. Diagn Mol Pathol 1997, 6:282-287 [DOI] [PubMed] [Google Scholar]
- 9.Wang YL, Bagg A, Pear W, Nowell PC, Hess JL: Chronic myelogenous leukemia: laboratory diagnosis and monitoring. Genes Chromosomes Cancer 2001, 32:97-111 [DOI] [PubMed] [Google Scholar]
- 10.Lee DS, Kim EC, Yoon BH, Kim WH, Yoon JH, Cho HI: Can minor bcr/abl translocation in acute leukemia be discriminated from major bcr/abl by extra-signal FISH analysis? Haematologica 2001, 86:991-992 [PubMed] [Google Scholar]
- 11.Grand FH, Chase A, Iqbal S, Nguyen DX, Lewis JL, Marley SB, Davidson RJ, Goldman JM, Gordon MY: A two-color BCR-ABL probe that greatly reduces the false positive and false negative rates for fluorescence in situ hybridization in chronic myeloid leukemia. Genes Chromosomes Cancer 1998, 23:109-115 [PubMed] [Google Scholar]
- 12.Dewald GW, Wyatt WA, Juneau AL, Carlson RO, Zinsmeister AR, Jalal SM, Spurbeck JL, Silver RT: Highly sensitive fluorescence in situ hybridization method to detect double BCR/ABL fusion and monitor response to therapy in chronic myeloid leukemia. Blood 1998, 91:3357-3365 [PubMed] [Google Scholar]
- 13.Buno I, Wyatt WA, Zinsmeister AR, Dietz-Band J, Silver RT, Dewald GW: A special fluorescent in situ hybridization technique to study peripheral blood and assess the effectiveness of interferon therapy in chronic myeloid leukemia. Blood 1998, 92:2315-2321 [PubMed] [Google Scholar]
- 14.Gleissner B, Rieder H, Thiel E, Fonatsch C, Janssen LA, Heinze B, Janssen JW, Schoch C, Goekbuget N, Maurer J, Hoelzer D, Bartram CR: Prospective BCR-ABL analysis by polymerase chain reaction (RT-PCR) in adult acute B-lineage lymphoblastic leukemia: reliability of RT-nested-PCR and comparison to cytogenetic data. Leukemia 2001, 15:1834-1840 [DOI] [PubMed] [Google Scholar]
- 15.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P: Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood 1995, 86:3118-3122 [PubMed] [Google Scholar]
- 16.Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV: The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood 1998, 92:3362-3367 [PubMed] [Google Scholar]
- 17.Yeoh E, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui C, Evans WE, Naeve C, Wong L, Downing JR: Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002, 1:133-143 [DOI] [PubMed] [Google Scholar]
- 18.Kurzrock R, Shtalrid M, Talpaz M, Kloetzer WS, Gutterman JU: Expression of c-abl in Philadelphia-positive acute myelogenous leukemia. Blood 1987, 70:1584-1588 [PubMed] [Google Scholar]
- 19.Kantarjian HM, O’Brien S, Smith TL, Cortes J, Giles FJ, Beran M, Pierce S, Huh Y, Andreeff M, Koller C, Ha CS, Keating MJ, Murphy S, Freireich EJ: Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. J Clin Oncol 2000, 18:547-561 [DOI] [PubMed] [Google Scholar]
- 20.Westbrook CA, Hooberman AL, Spino C, Dodge RK, Larson RA, Davey F, Wurster-Hill DH, Sobol RE, Schiffer C, Bloomfield CD: Clinical significance of the bcr-abl fusion gene in adult acute lymphoblastic leukemia: a cancer and leukemia group B study. Blood 1992, 80:2983-2990 [PubMed] [Google Scholar]
- 21.Mitterbauer G, Nemeth P, Wacha S, Cross NC, Schwarzinger I, Jaeger U, Geissler K, Greinix HT, Kalhs P, Lechner K, Mannhalter C: Quantification of minimal residual disease in patients with BCR-ABL-positive acute lymphoblastic leukaemia using quantitative competitive polymerase chain reaction. Br J Haematol 1999, 106:634-643 [DOI] [PubMed] [Google Scholar]
- 22.Legrand O, Perrot JY, Simonin G, Baudard M, Cadiou M, Blanc C, Ramond S, Viguie F, Marie JP, Zittoun R: Adult biphenotypic acute leukemia: an entity with poor prognosis which is related to unfavourable cytogenetics and P-glycoprotein over-expression. Br J Haematol 1998, 100:147-155 [DOI] [PubMed] [Google Scholar]
- 23.Gleissner B, Gokbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW, Fonatsch C, Heyll A, Voliotis D, Beck J, Lipp T, Munzert G, Maurer J, Hoelzer D, Thiel E: Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood 2002, 99:1536-1543 [DOI] [PubMed] [Google Scholar]
- 24.Crist W, Carroll A, Shuster J, Jackson J, Head D, Borowitz M, Behm F, Link M, Steuber P, Ragab A, Hirt A, Brock B, Land V, Pullen J: Philadelphia chromosome-positive childhood acute lymphoblastic leukemia: clinical and cytogenetic characteristics and treatment outcome: a Pediatric Oncology Group study. Blood 1990, 76:489-494 [PubMed] [Google Scholar]
- 25.Uckun FM, Nachman JB, Sather HN, Sensel MG, Kraft P, Steinherz PG, Lange B, Hutchinson R, Reaman GH, Gaynon PS, Heerema NA: Clinical significance of Philadelphia chromosome-positive pediatric acute lymphoblastic leukemia in context of contemporary intensive therapies: a report from the children’s cancer group. Cancer 1998, 83:2030-2039 [PubMed] [Google Scholar]
- 26.Schlieben S, Borkhardt A, Reinisch I, Ritterbach J, Janssen JW, Ratei R, Schrappe M, Repp R, Zimmermann M, Kabisch H, Janka-Schaub G, Bartram CR, Ludwig WD, Riehm H, Lampert F, Harbott J: Incidence and clinical outcome of children with BCR/ABL-positive acute lymphoblastic leukemia (ALL). A prospective RT-PCR study based on 673 patients enrolled in the German pediatric multicenter therapy trials ALL-BFM-90 and CoALL-05–92. Leukemia 1996, 10:957-963 [PubMed] [Google Scholar]
- 27.Mori T, Manabe A, Tsuchida M, Hanada R, Yabe H, Ohara A, Saito T, Nakazawa S: Allogeneic bone marrow transplantation in first remission rescues children with Philadelphia chromosome-positive acute lymphoblastic leukemia: Tokyo Children’s Cancer Study Group (TCCSG) studies L89–12 and L92–13. Med Pediatr Oncol 2001, 37:426-431 [DOI] [PubMed] [Google Scholar]
- 28.Carbonell F, Swansbury J, Min T, Matutes E, Farahat N, Buccheri V, Morilla R, Secker-Walker L, Catovsky D: Cytogenetic findings in acute biphenotypic leukemia. Leukemia 1996, 10:1283-1287 [PubMed] [Google Scholar]
- 29.Atlas of Genetics and Cytogenetics in Oncology and Haemotology. http://www.infobiogen.fr/services/chromcancer/Anomalies/t0922ANL.html, Feb 16, 2003
- 30.Schrappe M, Aricò M, Harbott J, Biondi A, Zimmermann M, Conter V, Reiter A, Valsecchi MG, Gadner H, Basso G, Bartram CR, Lampert F, Riehm H, Masera G: Philadelphia chromosome-positive (Ph+) childhood acute lymphoblastic leukemia: good initial steroid response allows early prediction of a favorable treatment outcome. Blood 1998, 92:2730-2741 [PubMed] [Google Scholar]
- 31.Lim LC, Heng KK, Vellupillai M, Tan LT, Boey BC, Lau LC, How GF: Molecular and phenotypic spectrum of de novo Philadelphia positive acute leukemia. Int J Mol Med 1999, 4:665-667 [DOI] [PubMed] [Google Scholar]
- 32.Boldt DH, Kopecky KJ, Head D, Gehly G, Radich JP, Appelbaum FR: Expression of myeloid antigens by blast cells in acute lymphoblastic leukemia of adults. The Southwest Oncology Group experience. Leukemia 1994, 8:2118-2126 [PubMed] [Google Scholar]
- 33.van Rhee F, Hochhaus A, Lin F, Melo JV, Goldman JM, Cross NC: p190 BCR-ABL mRNA is expressed at low levels in p210-positive chronic myeloid and acute lymphoblastic leukemias. Blood 1996, 87:5213-5217 [PubMed] [Google Scholar]
- 34.Kantarjian HM, Talpaz M, Dhingra K, Estey E, Keating MJ, Ku S, Trujillo J, Huh Y, Stass S, Kurzrock R: Significance of the P210 versus P190 molecular abnormalities in adults with Philadelphia chromosome-positive acute leukemia. Blood 1991, 78:2411-2418 [PubMed] [Google Scholar]
- 35.Secker-Walker LM, Craig JM: Prognostic implications of breakpoint and lineage heterogeneity in Philadelphia-positive acute lymphoblastic leukemia: a review. Leukemia 1993, 7:147-151 [PubMed] [Google Scholar]
- 36.Radich JP, Kopecky KJ, Boldt DH, Head D, Slovak ML, Babu R, Kirk J, Lee A, Kessler P, Appelbaum F, Gehly G: Detection of BCR-ABL fusion genes in adult acute lymphoblastic leukemia by the polymerase chain reaction. Leukemia 1994, 8:1688-1695 [PubMed] [Google Scholar]
- 37.Radich J, Gehly G, Lee A, Avery R, Bryant E, Edmands S, Gooley T, Kessler P, Kirk J, Ladne P, Thomas ED, Appelbaum FR: Detection of bcr-abl transcripts in Philadelphia chromosome-positive acute lymphoblastic leukemia after marrow transplantation. Blood 1997, 89:2602-2609 [PubMed] [Google Scholar]
- 38.Dombret H, Gabert J, Boiron J-M, Rigal-Huguet F, Blaise D, Thomas X, Delannoy A, Buzyn A, Bilhou-Babera C, Cayuela J-M, Fenaux P, Bourhis J-H, Fegeuex N, Charrin C, Boucheix C, Lheritier V, Esperou H, MacInyre E, Vernant J-P, Fiere D: Outcome of treatment in adults with Philadelphia chromosome-positive acute lymphoblastic leukemia-results of the prospective multicenter LALA-94 trial. Blood 2002, :2357-2366 [DOI] [PubMed] [Google Scholar]
- 39.Melo JV: The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood 1996, 88:2375-2384 [PubMed] [Google Scholar]
- 40.Saglio G, Pane F, Gottardi E, Frigeri F, Buonaiuto MR, Guerrasio A, de Micheli D, Parziale A, Fornaci MN, Martinelli G, Salvatore F: Consistent amounts of acute leukemia-associated P190BCR/ABL transcripts are expressed by chronic myelogenous leukemia patients at diagnosis. Blood 1996, 87:1075-1080 [PubMed] [Google Scholar]
- 41.Saglio G, Pane F, Martinelli G, Guerrasio A: BCR/ABL transcripts and leukemia phenotype: an unsolved puzzle. Leuk Lymphoma 1997, 26:281-286 [DOI] [PubMed] [Google Scholar]
- 42.Dhingra K, Talpaz M, Kantarjian H, Ku S, Rothberg J, Gutterman JU, Kurzrock R: Appearance of acute leukemia-associated P190 BCR-ABL in chronic myelogenous leukemia may correlate with disease progression. Leukemia 1991, 5:191-195 [PubMed] [Google Scholar]
- 43.Serrano J, Roman J, Sanchez J, Jimenez A, Castillejo JA, Herrera C, Gonzalez MG, Reina L, Rodriguez MC, Alvarez MA, Maldonado J, Torres A: Molecular analysis of lineage-specific chimerism and minimal residual disease by RT-PCR of p210 (BCR-ABL) and p190(BCR-ABL) after allogeneic bone marrow transplantation for chronic myeloid leukemia: increasing mixed myeloid chimerism and p190 (BCR-ABL) detection precedes cytogenetic relapse. Blood 2000, 95:2659-2665 [PubMed] [Google Scholar]
- 44.Onciu M, Bueso-Ramos C, Medeiros J, Ball G, Smith T, Lai R: Acute lymphoblastic leukemia in elderly patients: the Philadelphia chromosome may not be a significant adverse prognostic factor. Am J Clin Pathol 2002, 117:716-720 [DOI] [PubMed] [Google Scholar]
- 45.Killick S, Matutes E, Powles RL, Hamblin M, Swansbury J, Treleaven J, Zomas A, Atra A, Catovsky D: Outcome of biphenotypic acute leukemia. Haematologica 1999, 84:699-706 [PubMed] [Google Scholar]
- 46.Fletcher JA, Lynch EA, Kimball VM, Donnelly M, Tantravahi R, Sallan SE: Translocation (9;22) is associated with extremely poor prognosis in intensively treated children with acute lymphoblastic leukemia. Blood 1991, 77:435-439 [PubMed] [Google Scholar]
- 47.Friedmann AM, Weinstein HJ: The role of prognostic features in the treatment of childhood acute lymphoblastic leukemia. Oncologist 2000, 5:321-328 [DOI] [PubMed] [Google Scholar]
- 48.Thiebaut A, Vernant JP, Degos L, Huguet FR, Reiffers J, Sebban C, Lepage E, Thomas X, Fiere D: Adult acute lymphocytic leukemia study testing chemotherapy and autologous and allogeneic transplantation. A follow-up report of the French protocol LALA 87. Hematol Oncol Clin North Am 2000, 14:1353-1366 [DOI] [PubMed] [Google Scholar]
- 49.Mandelli F, Annino L, Rotoli B: The GIMEMA ALL 0183 trial: analysis of 10-year follow-up. Br J Haematol 1996, 92:665-672 [DOI] [PubMed] [Google Scholar]
- 50.Forman SJ, O’Donnell MR, Nademanee AP, Snyder DS, Bierman PJ, Schmidt GM, Fahey JL, Stein AS, Parker PM, Blume KG: Bone marrow transplantation for patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 1987, 70:587-588 [PubMed] [Google Scholar]
- 51.Snyder DS, Nademanee AP, O’Donnell MR, Parker PM, Stein AS, Margolin K, Somlo G, Molina A, Spielberger R, Kashyap A, Fung H, Slovak ML, Dagis A, Negrin RS, Amylon MD, Blume KG, Forman SJ: Long-term follow-up of 23 patients with Philadelphia chromosome-positive acute lymphoblastic leukemia treated with allogeneic bone marrow transplant in first complete remission. Leukemia 1999, 13:2053-2058 [DOI] [PubMed] [Google Scholar]
- 52.Kantarjian HM, Cortes J, O’Brien S, Giles FJ, Albitar M, Rios MB, Shan J, Faderl S, Garcia-Manero G, Thomas DA, Resta D, Talpaz M: Imatinib mesylate (STI571) therapy for Philadelphia chromosome-positive chronic myelogenous leukemia in blast phase. Blood 2002, 99:3547-3553 [DOI] [PubMed] [Google Scholar]
- 53.Sawyers CL, Hochhaus A, Feldman E, Goldman JM, Miller CB, Ottmann OG, Schiffer CA, Talpaz M, Guilhot F, Deininger MW, Fischer T, O’Brien SG, Stone RM, Gambacorti-Passerini CB, Russell NH, Reiffers JJ, Shea TC, Chapuis B, Coutre S, Tura S, Morra E, Larson RA, Saven A, Peschel C, Gratwohl A, Mandelli F, Ben-Am M, Gathmann I, Capdeville R, Paquette RL, Druker BJ: Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood 2002, 99:3530-3539 [DOI] [PubMed] [Google Scholar]
- 54.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL: Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001, 293:876-880 [DOI] [PubMed] [Google Scholar]
- 55.Hofmann WK, Jones LC, Lemp NA, de Vos S, Gschaidmeier H, Hoelzer D, Ottmann OG, Koeffler HP: Ph(+) acute lymphoblastic leukemia resistant to the tyrosine kinase inhibitor STI571 has a unique BCR-ABL gene mutation. Blood 2002, 99:1860-1862 [DOI] [PubMed] [Google Scholar]
- 56.Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K, Herrmann R, Lynch KP, Hughes TP: High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood 2002, 99:3472-3475 [DOI] [PubMed] [Google Scholar]
- 57.Verma A, Stock W: Management of adult acute lymphoblastic leukemia: moving toward a risk-adapted approach. Curr Opin Oncol 2001, 13:14-20 [DOI] [PubMed] [Google Scholar]
- 58.Sievers EL, Radich JP: Detection of minimal residual disease in acute leukemia. Curr Opin Hematol 2000, 7:212-216 [DOI] [PubMed] [Google Scholar]
- 59.Faderl S, Kantarjian HM, Talpaz M, Estrov Z: Clinical significance of minimal residual disease in leukemia. Int J Oncol 2000, 17:1277-1287 [DOI] [PubMed] [Google Scholar]
- 60.Cimino G, Elia L, Rapanotti MC, Sprovieri T, Mancini M, Cuneo A, Mecucci C, Fioritoni G, Carotenuto M, Morra E, Liso V, Annino L, Saglio G, De Rossi G, Foa R, Mandelli F: A prospective study of residual-disease monitoring of the ALL1/AF4 transcript in patients with t(4;11) acute lymphoblastic leukemia. Blood 2000, 95:96-101 [PubMed] [Google Scholar]
- 61.Pallisgaard N, Clausen N, Schroder H, Hokland P: Rapid and sensitive minimal residual disease detection in acute leukemia by quantitative real-time RT-PCR exemplified by t(12;21) TEL-AML1 fusion transcript. Genes Chromosomes Cancer 1999, 26:355-365 [DOI] [PubMed] [Google Scholar]
- 62.Saffroy R, Lemoine A, Brezillon P, Frenoy N, Delmas B, Goldschmidt E, Souleau B, Nedellec G, Debuire B: Real-time quantitation of bcr-abl transcripts in haematological malignancies. Eur J Haematol 2000, 65:258-266 [DOI] [PubMed] [Google Scholar]
- 63.Emig M, Saussele S, Wittor H, Weisser A, Reiter A, Willer A, Berger U, Hehlmann R, Cross NC, Hochhaus A: Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real-time quantitative RT-PCR. Leukemia 1999, 13:1825-1832 [DOI] [PubMed] [Google Scholar]
- 64.Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, Hehlmann R, Hiddemann W, Haferlach T: Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia 2002, 16:53-59 [DOI] [PubMed] [Google Scholar]
- 65.Eder M, Battmer K, Kafert S, Stucki A, Ganser A, Hertenstein B: Monitoring of BCR-ABL expression using real-time RT-PCR in CML after bone marrow or peripheral blood stem cell transplantation. Leukemia 1999, 13:1383-1389 [DOI] [PubMed] [Google Scholar]
- 66.Kreuzer KA, Lass U, Bohn A, Landt O, Schmidt CA: LightCycler technology for the quantitation of bcr/abl fusion transcripts. Cancer Res 1999, 59:3171-3174 [PubMed] [Google Scholar]
- 67.Amabile M, Giannini B, Testoni N, Montefusco V, Rosti G, Zardini C, Terragna C, Buonamici S, Ottaviani E, Soverini S, Fiacchini M, Bassi S, de Vivo A, Trabacchi E, Saglio G, Pane F, Baccarani M, Tura S, Martinelli G: Real-time quantification of different types of bcr-abl transcript in chronic myeloid leukemia. Haematologica 2001, 86:252-259 [PubMed] [Google Scholar]
- 68.Bolufer P, Sanz GF, Barragan E, Sanz MA, Cervera J, Lerma E, Senent L, Moreno I, Planelles MD: Rapid quantitative detection of BCR-ABL transcripts in chronic myeloid leukemia patients by real-time reverse transcriptase polymerase-chain reaction using fluorescently labeled probes. Haematologica 2000, 85:1248-1254 [PubMed] [Google Scholar]
- 69.Barbany G, Hagberg A, Olsson-Stromberg U, Simonsson B, Syvanen AC, Landegren U: Manifold-assisted reverse transcription-PCR with real-time detection for measurement of the BCR-ABL fusion transcript in chronic myeloid leukemia patients. Clin Chem 2000, 46:913-920 [PubMed] [Google Scholar]
- 70.Kreuzer KA, Lass U, Nagel S, Ellerbrok H, Pauli G, Pawlaczyk-Peter B, Siegert W, Huhn D, Schmidt CA: Applicability of an absolute quantitative procedure to monitor intra-individual bcr/abl transcript kinetics in clinical samples from chronic myelogenous leukemia patients. Int J Cancer 2000, 86:741-746 [DOI] [PubMed] [Google Scholar]
- 71.Stockschlader M, Hegewisch-Becker S, Kruger W, tom Dieck A, Mross K, Hoffknecht M, Berger C, Kohlschutter B, Martin H, Peters S, Kalisch H, Kuse R, Weh H, Zander A: Bone marrow transplantation for Philadelphia-chromosome-positive acute lymphoblastic leukemia. Bone Marrow Transplant 1995, 16:663-667 [PubMed] [Google Scholar]
- 72.Radich JP, Gooley T, Bryant E, Chauncey T, Clift R, Beppu L, Edmands S, Flowers ME, Kerkof K, Nelson R, Appelbaum FR: The significance of bcr-abl molecular detection in chronic myeloid leukemia patients “late,” 18 months or more after transplantation. Blood 2001, 98:1701-1707 [DOI] [PubMed] [Google Scholar]
- 73.Gehly GB, Bryant EM, Lee AM, Kidd PG, Thomas ED: Chimeric BCR-abl messenger RNA as a marker for minimal residual disease in patients transplanted for Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 1991, 78:458-465 [PubMed] [Google Scholar]
- 74.Miyamura K, Tanimoto M, Morishima Y, Horibe K, Yamamoto K, Akatsuka M, Kodera Y, Kojima S, Matsuyama K, Hirabayashi N: Detection of Philadelphia chromosome-positive acute lymphoblastic leukemia by polymerase chain reaction: possible eradication of minimal residual disease by marrow transplantation. Blood 1992, 79:1366-70 [PubMed] [Google Scholar]
- 75.Lin F, Goldman JM, Cross NC: A comparison of the sensitivity of blood and bone marrow for the detection of minimal residual disease in chronic myeloid leukaemia. Br J Haematol 1994, 86:683-685 [DOI] [PubMed] [Google Scholar]
- 76.Bock O, Lehmann U, Kreipe H: Quantitative intra-individual monitoring of BCR-ABL transcript levels in archival bone marrow trephines of patients with chronic myelogenous leukemia. J Mol Diagn 2003, 5:54-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsue K, Tabayashi T, Yamada K, Takeuchi M: Eradication of residual bcr-abl-positive clones by inducing graft-versus-host disease after allogeneic stem cell transplantation in patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Bone Marrow Transplant 2002, 29:63-66 [DOI] [PubMed] [Google Scholar]
- 78.Porter DL, Collins RH, Jr, Shpilberg O, Drobyski WR, Connors JM, Sproles A, Antin JH: Long-term follow-up of patients who achieved complete remission after donor leukocyte infusions. Biol Blood Marrow Transplant 1999, 5:253-261 [DOI] [PubMed] [Google Scholar]
- 79.Mughal TI, Yong A, Szydlo RM, Dazzi F, Olavarria E, vanRhee F, Kaeda J, Cross NCP, Craddock C, Kanfer E, Apperley J, Goldman JM: Molecular studies in patients with chronic myeloid leukemia in remission 5 years after allogeneic stem cell transplant define the risk of subsequent relapse. Br J Haematol 2001, 115:569-574 [DOI] [PubMed] [Google Scholar]
- 80.Bagg A: Chronic Myeloid Leukemia: a minimalist view of post-therapeutic monitoring. J Mol Diagn 2002, 4:1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lion T: Clinical implications of qualitative and quantitative polymerase chain reaction analysis in the monitoring of patients with chronic myelogenous leukemia: the European Investigators on Chronic Myeloid Leukemia Group. Bone Marrow Transplant 1994, 14:505-509 [PubMed] [Google Scholar]
- 82.Hochhaus A, Reiter A, Saussele S, Reichert A, Emig M, Kaeda J, Schultheis B, Berger U, Shepherd PC, Allan NC, Hehlmann R, Goldman JM, Cross NC: Molecular heterogeneity in complete cytogenetic responders after interferon-alpha therapy for chronic myelogenous leukemia: low levels of minimal residual disease are associated with continuing remission. Blood 2000, 95:62-66 [PubMed] [Google Scholar]
- 83.Lin F, vanRhee F, Goldman JM, Cross NC: Kinetics of increasing BCR-ABL transcript numbers in chronic myeloid leukemia patients who relapse after bone marrow transplantation. Blood 1996, 87:4473-4478 [PubMed] [Google Scholar]
- 84.Olavarria E, Kanfer E, Szydlo R, Kaeda J, Rezvani K, Cwynarski K, Pocock C, Dazzi F, Craddock C, Apperley JF, Cross NCP, Goldman JM: Early Detection of BCR-ABL transcripts by quantitative reverse transcriptase-polymerase chain reaction predicts outcome after allogeneic stem cell transplantation for chronic myeloid leukemia. Blood 2001, 97:1560-1565 [DOI] [PubMed] [Google Scholar]
- 85.National Comprehensive Cancer Network Practice Guidelines for Chronic Myelogenous Leukemia. Version 1.2002;10/30/01