Figure 2.
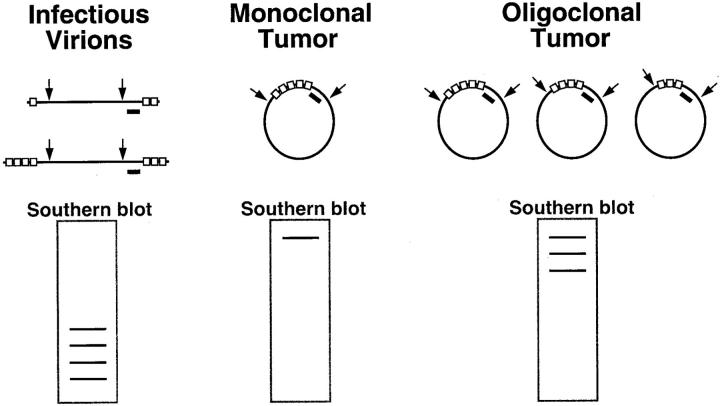
The EBV clonality assay evaluates clonality with respect to the structure of the EBV genome. The assay is based on the presence of variable numbers of tandem repeat sequences (shown as open boxes) at the ends of the linear viral genome. On infection of a cell, these ends join to form an episome by fusing up to 20 terminal repeat sequences. When an infected cell undergoes malignant transformation, the same fused terminal repeat structure is inherited by all progeny of the malignant clone. The clonality assay is accomplished by Southern blot analysis of DNA extracted from patient tissue and digested with BamHI restriction endonuclease (shown by arrows) to cut the EBV genome at sites flanking the terminal repeats. This results in restriction fragments that are recognized by a DNA probe (black bar). Examination of the band pattern on Southern blots reveals that infectious virions produce a ladder array of small bands. In contrast, a monoclonal tumor exhibits a single band of high molecular weight, and an oligoclonal tumor has several such bands.