Abstract
Histopathological differentiation between hepatocellular adenoma and well differentiated hepatocellular carcinoma (HCC) may be a difficult task in small biopsies and occasionally in resected tumor specimens. Whether the analysis of chromosome aberrations can contribute to a more precise discrimination has not been analyzed systematically up to now. Therefore, fluorescence in situ hybridization was applied to 28 cases of adenoma and well differentiated carcinoma, using centromeric probes for chromosomes 1, 6, 7, 8, and X. None of 14 adenomas revealed an aberrant count in the analyses performed. By contrast, 13/14 carcinomas demonstrated aberrations for 2–5 chromosomes/case. Chromosome 1 was aberrant in 8/12 cases informative for this probe (67%), chromosomes 6 and 7 were aberrant in 9/14 cases (64%), chromosome 8 was aberrant in 11/14 cases (79%), and chromosome X in 7/14 cases (50%). Taking results for chromosomes 1 and 8 together, 13/14 HCC revealed aberrations for at least one of these chromosomes. Probes for 6, 7, and X revealed no additional aberrant cases.Thus, FISH for chromosomes 1 and 8, extended by probes for chromosomes 6, 7 and X, represents a promising approach toward a more accurate differentiation between hepatocellular adenoma and carcinoma.
Hepatocellular carcinoma (HCC) represents the most frequent malignant tumor of the liver 1 and is associated with different etiologies such as viral infection and toxic agents. 2 Despite the advances in sonographic and radiological techniques, histological examination still remains the gold standard in the diagnosis of HCC. 3 Whereas the identification of moderate and poorly differentiated HCC is easily achieved by histopathology, identification of well differentiated HCC is more difficult. Distinction from liver cell adenoma still remains a diagnostic challenge, particularly in small biopsies. 4, 5
Analysis of cytogenetic aberrations in HCC could provide a potential solution to problematic histological queries. Conventional cytogenetics (CG) is not useful for this purpose due to well-known difficulties in obtaining metaphases necessary for karyotyping. 6, 7, 8, 9 The alternative to CG, comparative genomic hybridization (CGH), permits karyotyping without the need for metaphase preparation. 10 With this technique, larger numbers of HCC have been analyzed, revealing typical aberration patterns not only in moderate or poorly differentiated HCC but also in well-differentiated samples. 11, 12, 13 These patterns with numerous aberrations were strikingly different from the low number of aberrations detected in a CGH study analyzing hepatocellular adenoma (HCA). 14 However, CGH is based on an elaborate and time-consuming procedure comparable to conventional CG and is difficult to apply in small biopsies in daily routine.
In comparison to CGH, fluorescence in situ hybridization (FISH) detects aberrations of defined chromosome loci in intact nuclei and preserved histological architecture even in small specimens. Since the main aberrations occurring in HCC are now known, based on the previous CGH results, it seems appropriate to analyze HCC and HCA by FISH, taking probes for those loci most often affected. For this purpose, we analyzed histological samples of 28 cases of HCA and well-differentiated HCC by FISH with a panel of 5 centromere-specific probes.
Materials and Methods
Tissue samples from 28 patients at the Medizinische Hochschule Hannover were analyzed. There were 14 patients in the group suffering from HCA, 11 of them female and 3 male (Table 1) . Ages ranged from 27 to 59 years with a mean of 37 years, as listed in Table 1 . Fourteen patients were known to have a well-differentiated HCC; 4 were female and 10 were male. Ages ranged from 41 to 76 years with a mean of 63 years. Diagnosis of HCA and HCC was based on hematoxylin and eosin (H&E)-stained sections and also periodic acid-Schiff, Elastica van Gieson, Orcein, and iron stains. To ensure accuracy of histological diagnoses, HCA was assumed only when disease-free survival of the patients exceeded 4 years. Furthermore, HCA samples were included only when the tumor was resected and analyzed by multiple additional tissue samples. To confirm diagnoses of HCC, samples were accepted when histological examination of the tumor was possible, as in HCA. In patients not undergoing surgical resection, HCC was assumed when obvious signs of malignancy, in particular metastases, were detectable.
Table 1.
Age and Gender of the Patients Analyzed
| Patients with adenomas | Patients with carcinomas | ||||
|---|---|---|---|---|---|
| Gender | Age | Gender | Age | ||
| HCA 1 | w | 30 | HCC 1 | m | 71 |
| HCA 2 | w | 30 | HCC 2 | m | 61 |
| HCA 3 | w | 48 | HCC 3 | m | 64 |
| HCA 4 | w | 33 | HCC 4 | m | 48 |
| HCA 5 | w | 27 | HCC 5 | m | 67 |
| HCA 6 | w | 27 | HCC 6 | m | 76 |
| HCA 7 | w | 27 | HCC 7 | w | 75 |
| HCA 8 | m | 43 | HCC 8 | m | 64 |
| HCA 9 | m | 59 | HCC 9 | w | 74 |
| HCA 10 | w | 47 | HCC 10 | w | 64 |
| HCA 11 | m | 38 | HCC 11 | m | 72 |
| HCA 12 | w | 33 | HCC 12 | m | 41 |
| HCA 13 | w | 37 | HCC 13 | m | 48 |
| HCA 14 | w | 34 | HCC 14 | w | 59 |
FISH analyses were performed either on biopsies taken in vivo by fine needle aspiration (n = 15) or on biopsies taken from tissue obtained after surgical removal of the tumors (n = 13). Specimens were fixed for at least 24 hours in formalin, embedded in paraffin, and sampled together in a multi-tissue block (MultiBlock; Zytomed, Berlin, Germany).
FISH
FISH was performed on all samples with centromere-specific probes. Centromeric regions contain highly repetitive sequences, that are much easier to detect with FISH probes than low- or even single-copy sequences of chromosomes. The probes used were chosen with regard to CGH results in well differentiated HCC detected by our own group and other authors. In these studies, the most often altered chromosomes included 1, 4, 5, 6, 7, 8, 16, 17, and X with centromeric regions frequently affected in chromosomes 1, 4, 6, 7, 8, and X. Because a centromeric probe for chromosome 4 was not commercially available at the time the experimental part of the study was performed, this chromosome was excluded. The panel of probes applied to the specimens therefore included centromeric probes for 1, 6, 7, 8, and X (all Oncor, Heidelberg, Germany).
FISH was performed on 5-μm sections mounted on superfrost slides (Omnilab, Gehrten, Germany). Tissue sections were baked overnight at 56°C and then deparaffinized by immersing in xylene for 20 minutes and then in graded ethanol. Slides were placed in citric acid solution (0.01 mol/L) and heated in a microwave oven at 900 and 600 W for 15 minutes each. Diluted RNase A (0.1%) was added to the sections for 10 minutes and then rinsed in PBD (Oncor). Incubation with 3% H2O2 for 10 minutes was carried out at room temperature, followed by washing in PBD. Afterward, slides were washed in graded ethanol and air-dried for 5 minutes. A total of 0.5 μl of the probe was added to 10 μl Hybrisol VI (Oncor) and pipetted onto the slide, placed under a glass coverslip, sealed with rubber cement, heated to 92°C for 12 minutes, and incubated overnight at 37°C in a humifidied chamber. Detection started with rinsing the sections in 0.25× SSC at 60°C for 5 minutes followed by a short wash in PBD. Then 30 μl of horseradish peroxidase (HRP), diluted 1:30, were added for 20 minutes at 37°C under a coverslip. Washing in PBD followed. Thirty microliters of fluorescein isothiocyanate-conjugated tyramine (DuPont NEN, Boston, MA) were added and incubated for 20 minutes at 37°C. After rinsing in PBD, counterstaining was done with 5 μl of propidium iodide (Oncor), and the slides were placed under a coverslip.
Evaluation of signals was done using an epifluorescence microscope (Axiophot; Zeiss, Oberkochen, Germany) equipped with a fluorescein/rhodamine filter set and a 50 W mercury lamp. Only bright signals not connected to a second signal were counted. At least 100 nuclei were evaluated in each case.
Results
In a first step, signal distribution was determined in non-neoplastic liver tissues. For this purpose, 55 specimens of normal liver were analyzed. As listed in Table 2 , the mean values for distribution of signals were 81% of nuclei bearing two signals, 17% revealing one signal, and 2.3% showing three or more signals. The standard deviations were 4.7, 4.2, and 2.6%, respectively. Pursuant to the recommendations of Ward et al, 15 three standard deviations were added to the mean values and defined as monosomy and trisomy at 29% and 10%, respectively.
Table 2.
Mean, Standard Deviation (SD), and Threshold for Defining Monosomy and Polysomy Based on the Evaluation of 55 Normal Specimens
| Value | Signals/nucleus (%) | ||
|---|---|---|---|
| 1 | 2 | 3 | |
| Mean | 17 | 81 | 2.3 |
| SD | 4.2 | 4.7 | 2.6 |
| Threshold* | 29 | 95 | 10 |
Obtained by adding 3 × SD to the mean.
Hepatocellular Adenoma
One signal was seen in 4.3 to 24.6% of cells analyzed. Two signals per nucleus were seen in 73.8 to 91.1% of nuclei (Table 3 , Figure 1 ). Three or more signals were detectable in 0.0 to 7.5% of the cells. None of these values passed the thresholds of 29% and 10% defining monosomy and trisomy, respectively. In seven analyses of seven different cases, an analysis was not informative due to lack of specific probe binding.
Table 3.
Results of FISH for the Adenomas and Carcinomas Analyzed
| Adenomas | Carcinomas | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Probe | Signals/nucleus (%) | Patient | Probe | Signals/nucleus (%) | ||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||||
| HCA 1 | cen1 | 8.9 | 91.1 | 0.0 | HCC 1 | cen1 | 32 | 66.2 | 1.8 |
| cen6 | 11.2 | 88.1 | 0.7 | cen6 | 26.8 | 70.8 | 2.4 | ||
| cen7 | 13.3 | 86.7 | 0.0 | cen7 | 30.3 | 69.3 | 0.4 | ||
| cen8 | 23.6 | 76.4 | 0.0 | cen8 | 19.4 | 78.2 | 2.4 | ||
| cenX | 14.8 | 85.2 | 0.0 | cenX | 79 | 21 | 0 | ||
| HCA 2 | cen1 | 22.7 | 75.4 | 2.0 | HCC 2 | cen1 | 17.5 | 68.8 | 13.8 |
| cen6 | 18.6 | 79.5 | 1.9 | cen6 | 24.7 | 65.4 | 9.9 | ||
| cen7 | 18.4 | 78.9 | 2.6 | cen7 | 27.4 | 54.8 | 17.8 | ||
| cen8 | 17.8 | 75.6 | 6.7 | cen8 | 19.1 | 66.7 | 14.2 | ||
| cenX | 17.9 | 79.5 | 2.7 | cenX | 94.4 | 5.6 | 0 | ||
| HCA 3 | cen1 | n.s. | HCC 3 | cen1 | 12.9 | 53.6 | 33.6 | ||
| cen6 | 17.5 | 80.3 | 2.2 | cen6 | 21.7 | 77.1 | 1.2 | ||
| cen7 | 20.4 | 76.9 | 2.8 | cen7 | 14.9 | 79.9 | 5.2 | ||
| cen8 | 14.1 | 82.0 | 3.9 | cen8 | 17.6 | 67.6 | 14.7 | ||
| cenX | 10.0 | 87.2 | 2.8 | cenX | 93.8 | 6.3 | 0 | ||
| HCA 4 | cen1 | 21.3 | 78.1 | 0.6 | HCC 4 | cen1 | 21 | 77.4 | 1.6 |
| cen6 | n.s. | cen6 | 13.5 | 58.5 | 28 | ||||
| cen7 | 12.2 | 86.0 | 1.8 | cen7 | 19.2 | 53.7 | 27.1 | ||
| cen8 | 12.6 | 79.9 | 7.5 | cen8 | 14.3 | 56 | 29.7 | ||
| cenX | 7.4 | 90.9 | 1.7 | cenX | 40.2 | 52.1 | 7.7 | ||
| HCA 5 | cen1 | 21.0 | 78.1 | 1.0 | HCC 5 | cen1 | 47.4 | 52.6 | 0 |
| cen6 | 18.7 | 79.4 | 1.9 | cen6 | 41.5 | 58.5 | 0 | ||
| cen7 | 14.7 | 85.3 | 0.0 | cen7 | 31.4 | 66.7 | 2 | ||
| cen8 | 15.2 | 83.7 | 1.1 | cen8 | 42.7 | 56.3 | 1 | ||
| cenX | 11.6 | 87.9 | 0.5 | cenX | 96.6 | 3.4 | 0 | ||
| HCA 6 | cen1 | 17.6 | 79.1 | 3.3 | HCC 6 | cen1 | 22.3 | 64.5 | 13.2 |
| cen6 | 16.1 | 82.3 | 1.6 | cen6 | 11.1 | 49.3 | 39.6 | ||
| cen7 | 11.2 | 84.6 | 4.2 | cen7 | 24.2 | 75.8 | 0 | ||
| cen8 | 16.7 | 78.5 | 4.9 | cen8 | 10.6 | 64.2 | 25.1 | ||
| cenX | 12.7 | 83.7 | 3.6 | cenX | 95.7 | 4.3 | 0 | ||
| HCA 7 | cen1 | 7.0 | 93.0 | 0.0 | HCC 7 | cen1 | 7.8 | 34.6 | 57.5 |
| cen6 | 18.1 | 78.5 | 3.5 | cen6 | 9 | 16.5 | 74.4 | ||
| cen7 | 15.4 | 84.6 | 0.0 | cen7 | 6.5 | 37.4 | 56.1 | ||
| cen8 | 21.4 | 77.1 | 1.5 | cen8 | 7.5 | 39.7 | 52.7 | ||
| cenX | 7.0 | 91.5 | 1.5 | cenX | 5 | 35 | 60 | ||
| HCA 8 | cen1 | 24.2 | 75.8 | 0.0 | HCC 8 | cen1 | 27.4 | 72.1 | 0.5 |
| cen6 | 12.8 | 85.0 | 2.3 | cen6 | 19.3 | 54.8 | 25.9 | ||
| cen7 | 17.0 | 82.5 | 0.6 | cen7 | 8.5 | 79.7 | 11.9 | ||
| cen8 | 25.0 | 73.3 | 1.7 | cen8 | 76.2 | 20.5 | 3.3 | ||
| cenX | 98.4 | 1.6 | 0.0 | cenX | 31 | 68.2 | 0.8 | ||
| HCA 9 | cen1 | 17.1 | 82.9 | 0.0 | HCC 9 | cen1 | n.s. | ||
| cen6 | 17.4 | 81.4 | 1.2 | cen6 | 8.3 | 78.3 | 13.4 | ||
| cen7 | 12.7 | 85.7 | 1.6 | cen7 | 7.9 | 68.8 | 23.3 | ||
| cen8 | n.s. | cen8 | 17.5 | 72.1 | 10.4 | ||||
| cenX | 98.4 | 1.6 | 0.0 | cenX | 8.9 | 78.9 | 12.2 | ||
| HCA 10 | cen1 | 22.0 | 78.0 | 0.0 | HCC 10 | cen1 | 4.7 | 49.1 | 46.2 |
| cen6 | 4.3 | 92.8 | 2.9 | cen6 | 93 | 7 | 0 | ||
| cen7 | 13.2 | 85.8 | 0.9 | cen7 | 5.3 | 31.6 | 63.2 | ||
| cen8 | 10.3 | 89.7 | 0.0 | cen8 | 19 | 77.1 | 3.8 | ||
| cenX | 10.7 | 88.4 | 0.8 | cenX | 2.7 | 33.3 | 64 | ||
| HCA 11 | cen1 | 22.7 | 77.3 | 0.0 | HCC 11 | cen1 | 12.4 | 83.3 | 4.3 |
| cen6 | n.s. | cen6 | 34.7 | 65.3 | 0 | ||||
| cen7 | 16.7 | 82.1 | 1.2 | cen7 | 22.4 | 70.6 | 7 | ||
| cen8 | 11.5 | 86.9 | 1.6 | cen8 | 1.7 | 30.9 | 67.4 | ||
| cenX | 14.4 | 84.0 | 1.6 | cenX | 79.2 | 20.8 | 0 | ||
| HCA 12 | cen1 | 17.7 | 82.3 | 0.0 | HCC 12 | cen1 | 24.6 | 38.6 | 36.8 |
| cen6 | 9.1 | 89.3 | 1.7 | cen6 | 11.4 | 33.5 | 55.1 | ||
| cen7 | 12.3 | 87.7 | 0.0 | cen7 | 11.8 | 36.8 | 51.3 | ||
| cen8 | 8.6 | 91.4 | 0.0 | cen8 | 9.2 | 20.2 | 70.5 | ||
| cenX | n.s. | cenX | 27.7 | 72.3 | 0 | ||||
| HCA 13 | cen1 | 11.7 | 85.4 | 2.9 | HCC 13 | cen1 | 13.5 | 55.8 | 30.8 |
| cen6 | 18.9 | 81.1 | 0.0 | cen6 | 9.5 | 47.8 | 42.7 | ||
| cen7 | 14.7 | 84.5 | 0.9 | cen7 | 6.1 | 59.2 | 34.7 | ||
| cen8 | 24.6 | 73.8 | 1.5 | cen8 | 7 | 50.9 | 42.1 | ||
| cenX | n.s. | cenX | 40.1 | 56.6 | 3.3 | ||||
| HCA 14 | cen1 | 12.6 | 85.8 | 1.6 | HCC 14 | cen1 | n.s. | ||
| cen6 | 16.0 | 82.4 | 1.7 | cen6 | 24.9 | 70.7 | 4.4 | ||
| cen7 | 11.7 | 84.5 | 3.9 | cen7 | 19.4 | 79.8 | 0.8 | ||
| cen8 | 21.4 | 78.6 | 0.0 | cen8 | 20.6 | 78.5 | 1 | ||
| cenX | n.s. | cenX | 18.3 | 78 | 3.7 | ||||
The counts found abnormal in the sense of monosomy or polysomy are printed in bold. Aberrant counts were seen in carcinomas only.
n.s., not successfully performed.
Figure 1.
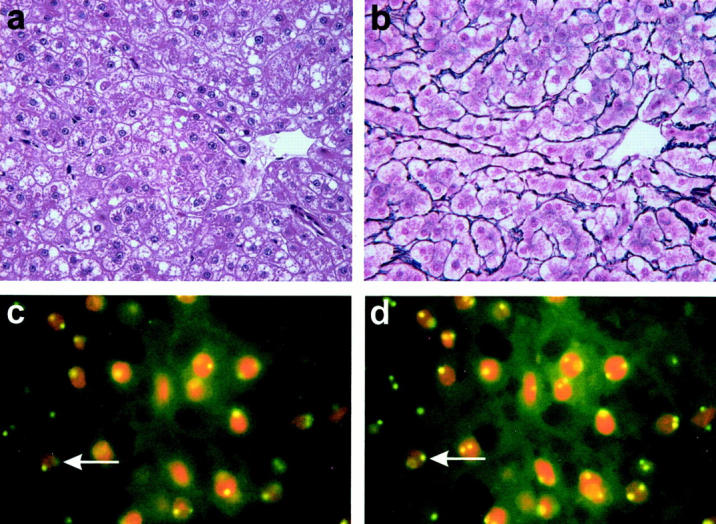
H&E-stained section of case HCA 12 reveals variations in nuclear size and prominent nucleoli (a; original magnification, ×400). Silver stain demonstrates a preserved delicate network of reticulin fibers around the tumor cells (b; ×400). FISH for chromosome 6 gives a normal distribution of signals, as indicated by one or two fluorescent signals in the nuclei (c; ×1000). Due to cutting artifacts, not all nuclei are expected to bear two signals. The number of signals shown in this photograph, however, is too low, and varying the plane of focus for approximately 2 μm brings up additional signals, as indicated for an exemplary cell by an arrow (d; ×1000).
Well-Differentiated Hepatocellular Carcinoma
One signal was seen in 1.7% to 76.2% of cells, two signals were seen in 20.2% to 79.8%, and three or more signals occurred in 0% to 70.5% (Table 3 , Figure 2 ). Chromosome 1 was found to be aberrant in 8 samples, chromosome 6 in 9 samples, chromosome 7 in 10 samples, chromosome 8 in 11, and chromosome X in 8 samples. Five chromosomes were found to be aberrant in 2 cases, 4 chromosomes were aberrant in 4 samples, 3 chromosomes were aberrant in 6 samples, 2 chromosomes were aberrant in 1 case, and in 1 sample no aberrant count was seen (Table 4) . In two analyses in two different cases, experiments were not informative.
Figure 2.
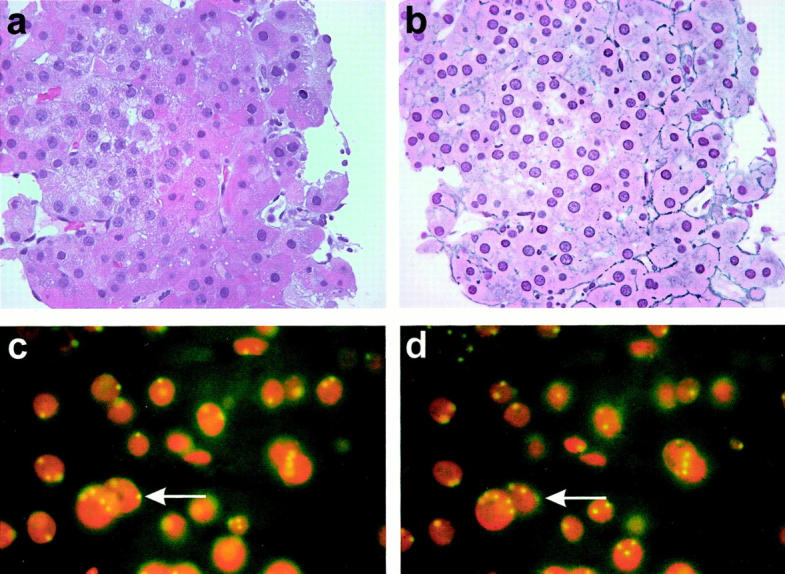
Variations of nuclear size and prominent nucleoli are also seen in case HCC 7 (a; original magnification, ×400). Reticulin fibers are seen in a part of the tumor, whereas in some areas the network is interrupted (b; ×400). FISH for chromosome 8 demonstrates polyploidy for this chromosome (c; ×1000). Varying the plane of focus again brings up additional signals (d; ×1000).
Table 4A.
Number of Aberrant Chromosomes in the HCC Samples
| Patient | No. of aberrant chromosomes |
|---|---|
| HCC 1 | 3 |
| HCC 2 | 3 |
| HCC 3 | 2 |
| HCC 4 | 3 |
| HCC 5 | 4 |
| HCC 6 | 3 |
| HCC 7 | 5 |
| HCC 8 | 4 |
| HCC 9 | 4 |
| HCC 10 | 3 |
| HCC 11 | 3 |
| HCC 12 | 4 |
| HCC 13 | 5 |
| HCC 14 | 0 |
Table 4B.
HCC Cases Found Aberrant for Particular Chromosomes
| Chromosome | Number of cases aberrant for this chromosome | Number of cases informative for this chromosome |
|---|---|---|
| 1 | 8 (67) | 12 |
| 6 | 9 (64) | 14 |
| 7 | 9 (64) | 14 |
| 8 | 11 (79) | 14 |
| X | 7 (50) | 14 |
Statistical Evaluation
The number of aberrations found in HCA and HCC samples was significantly different (P < 0.01, Mann-Whitney U test). Specificity of FISH in detecting HCC was 100%, and sensitivity was 92.3%. Positive predictive value was 100%, and negative predictive value reached 93.3%.
Discussion
Knowledge about cytogenetic alterations in HCC has increased over the past few years due to the application of new molecular techniques such as CGH. Larger numbers of HCC have now been analyzed, and recurrent patterns of chromosomal imbalances have been identified. 11, 12, 13 In particular, imbalances of chromosomes 1, 4, 6, 7, 8, and X, including total and partial gains and losses, have been demonstrated. Although not all of these aberrations were detectable in every case analyzed, at least some of them were found in varying combinations in all HCC cases described. By contrast, HCA demonstrated a much lower number and different chromosomes affected compared to HCC. 14 Based on these findings we used FISH as an alternative method to CGH to analyze HCA and HCC. The main reason for this approach was that FISH is easier to perform and much easier to evaluate than CGH. Whereas CGH requires a karyotype analysis similar to conventional cytogenetics, FISH requires only the counting of single signal spots in the nuclei. Therefore, the correct identification of chromosomes, which requires a lot of experience, is not mandatory in FISH.
Obvious differences in signal distributions of the particular chromosomes were seen between HCA and HCC. Whereas none of the HCA revealed aberrant counts for any of the chromosomes analyzed, HCC demonstrated aberrations for at least two chromosomes in 13/14 cases. In one HCC sample, probes for chromosomes 6, 7, 8, and X demonstrated normal distribution of signals and the probe for chromosome 1 did not give sufficient results. In this patient, partial hepatectomy was performed after diagnosis of HCC and the tumor was analyzed by CGH. Beside aberrations for 5q, 6p, 8p, 17p, and 17q, a gain of 8q was detectable, confirming a trisomic count seen in FISH using a locus-specific probe for 8q21. However, this probe failed to give reliable results in many cases examined. Therefore, it cannot be recommended for routine purposes and was consequently excluded from this study.
The restriction to five centromeric probes for FISH analysis is based on aspects of availability and applicability in daily routine. Centromeric probes most often give brighter signals than probes localized on the arms of the chromosomes. Evaluation of the signals can be done by epifluorescence microscopy with a standard filter set without the need for sophisticated technical equipment. All centromeric probes as well as the detection system are commercially available and can be processed by the same protocol. For these reasons, chromosome 4, also often reported to be altered in HCC, 11, 12, 16 was not analyzed in this study, since a digoxigenin-labeled probe was not available at the time our study was performed. This does not seem to hamper FISH analysis, since results obtained for chromosomes 1 and 8 already highlighted 13/14 cases as aberrant. No additional case was detected by analysis of chromosomes 6,7, and X. Therefore, it may be useful to apply probes for chromosomes 1 and 8 at first and, if one or both of these probes fail to give any results, to add the probes specific for chromosomes 6, 7, and X.
The aneuploidy found by the panel of probes is seen not only in HCC but also in a variety of other malignant tumors affecting the same chromosomes in similar patterns, as summarized by Mitelman et al. 17 Lengauer et al 18 discussed these findings as an increased genetic instability, based on the inability of the aberrant cell to control chromosomal alterations. This assumption is underscored by the observation that the chromosome changes found in distinct carcinomas are not always identical for all chromosomes. The mechanisms responsible for this genetic instability are not yet known and require further investigation. Nevertheless, they are diagnostically helpful, as shown here for HCA and HCC.
In conclusion, FISH for chromosomes 1 and 8, extended by probes specific for chromosomes 6, 7, and X, enables the differentiation of HCA and well-differentiated HCC to be performed with a high degree of accuracy. Particularly with regard to small biopsies, FISH represents a promising adjunct to classical histology in the differentiation between hepatocellular adenoma and carcinoma.
Address reprint requests to Dr. Ludwig Wilkens, Institut für Pathologie der Medizinischen Hochschule Hannover, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany. E-mail: wilkens.ludwig@mh-hannover.de.
References
- 1.Bosch F, Munoz N: Hepatocellular carcinoma in the world: epidemiologic questions. Adv Appl Biotechnol 1991, 13:55-56 [Google Scholar]
- 2.Wogan GN: Aflatoxins as risk factors for hepatocellular carcinoma in humans. Cancer Res 1992, 52:2114-2118 [PubMed] [Google Scholar]
- 3.Ishak KG, Anthony PP, Sobin LH: Histological Typing of Tumours of the Liver, 2nd ed. 1998, Springer, New York
- 4.Carrasco D, Prieto M, Pallardo L, Moll JL, Cruz JM, Munoz C, Berenguer J: Multiple hepatic adenomas after long-term therapy with testosterone enanthate: review of the literature. Hepatology 1985, 1:573-578 [DOI] [PubMed] [Google Scholar]
- 5.: Liver Cancer Study Group of Japan: The General Rules for the Clinical and Pathological Study of Primary Liver, 2nd ed. 1987, :pp 25-26 Kanehara Press, Tokyo [Google Scholar]
- 6.Simon D, Knowles BB, Weith A: Abnormalities of chromosome 1 and loss of heterozygosity on 1p in primary hepatomas. Oncogene 1991, 6:765-770 [PubMed] [Google Scholar]
- 7.Saito H, Morizane T, Watanabe T, Kagawa T, Iwabuchi MN, Kumagai N, Inagaki Y, Tsuchimoto K, Tsuchiya M: Establishment of a human cell line (HCC-T) from a patient with hepatoma bearing no evidence of hepatitis B or A virus infection. Cancer 1989, 64:1054-1060 [DOI] [PubMed] [Google Scholar]
- 8.Chen HL, Chen YC, Chen DS: Chromosome 1p aberrations are frequent in human primary hepatocellular carcinoma. Cancer Genet Cytogenet 1996, 86:102-106 [DOI] [PubMed] [Google Scholar]
- 9.Bardi G, Johansson B, Pandis N, Heim S, Mandahl N, Andren SA, Hagerstrand I, Mitelman F: Cytogenetic findings in three primary hepatocellular carcinomas. Cancer Genet Cytogenet 1992, 58:191-195 [DOI] [PubMed] [Google Scholar]
- 10.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 11.Wong N, Lai P, Lee S-W, Fan S, Pang E, Liew C-T, Sheng Z, Lau J, Johnson PJ: Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis. Am J Pathol 1999, 154:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusano N, Shirashi K, Kubo K, Oga A, Okita K, Sasaki K: Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology 1999, 29:1858-1862 [DOI] [PubMed] [Google Scholar]
- 13.Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu N: Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology 1999, 29:1208-1214 [DOI] [PubMed] [Google Scholar]
- 14.Wilkens L, Bredt M, Flemming P, Becker T, Kubicka S, Kreipe H: Differentiation of liver cell adenomas from well differentiated hepatocellular carcinomas by comparative genomic hybridisation. J Pathol 2001, 193:476-482 [DOI] [PubMed] [Google Scholar]
- 15.Ward BE, Gersen SL, Carelli MP, McGuire NM, Dackowski WR, Weinstein M, Sandlin C, Warren R, Klinger KW: Rapid prenatal diagnosis of chromosomal aneuploidies by fluorescence in situ hybridization: clinical experience with 4,500 specimens. Am J Hum Genet 1993, 52:854-865 [PMC free article] [PubMed] [Google Scholar]
- 16.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A: Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 1997, 18:59-65 [PubMed] [Google Scholar]
- 17.Mitelman F, Mertens F, Johansson B: A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet 1997, 15:417-474 [DOI] [PubMed] [Google Scholar]
- 18.Lengauer CL, Kinzler KW, Vogelstein B: Genetic instabilities in human cancer. Nature 1998, 396:643-649 [DOI] [PubMed] [Google Scholar]


