Figure 1.
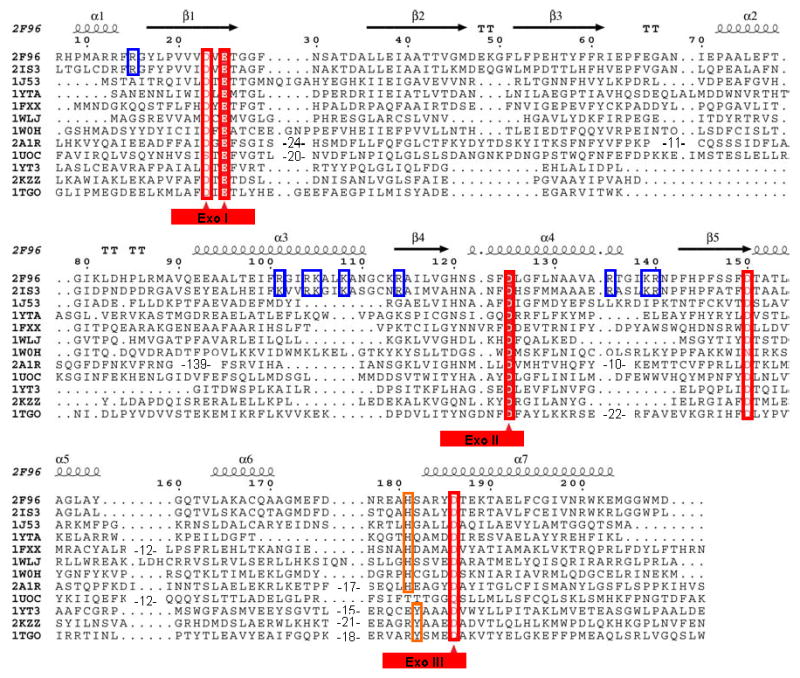
Structure based multiple alignment of E. coli and P. aeruginosa RNase T and related nucleases. Structures included here are: E. coli RNase T (PDB ID: 2IS3); P. aeruginosa putative RNase T (PDB ID: 2F96); E. coli DNA polymerase III ε subunit (PDB ID: 1J53); E. coli Oligoribonuclease (PDB ID: 1YTA); E. coli exonuclease I (PDB ID: 1FXX); three human 3′-5′ exoribonucleases, PARN (PDB ID: 2A1R), 3′hExo (PDB ID: 1W0H) and ISG20 (PDB ID: 1WLJ); yeast POP2 protein exonuclease domain (PDB ID: 1UOC); E. coli RNase D (PDB ID: 1YT3); Klenow fragment (PDB ID: 2KZZ); and T. gorgonarius DNA polymerase (PDB ID: 1TGO). The last three belong to the DEDDy subgroup, while the others have the DEDDh folds. Sequence alignments were initially generated using Tcoffee (http://www.tcoffee.org) (Notredame et al., 2000), followed by some manual adjustment. The three conserved Exo motifs are labeled at bottom with the DEDD residues marked by red triangles. The NBS basic residues conserved in RNase T orthologs are highlighted using blue rectangles. The numbering on the top is based on the E. coli RNase T sequence, with the secondary structure of P. aeruginosa RNase T.
