Figure 5.
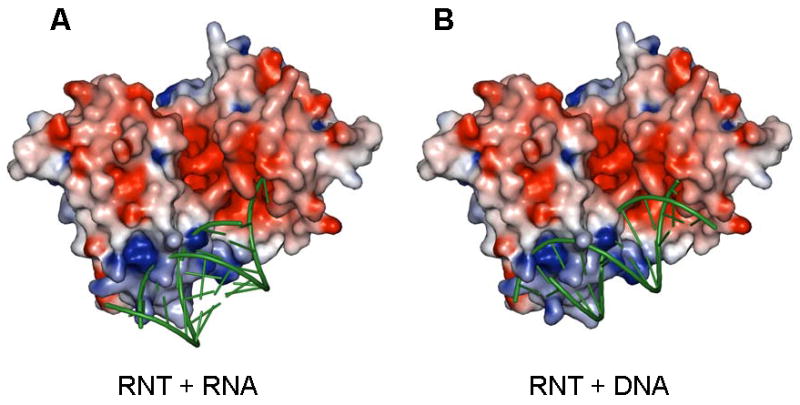
Possible binding modes for duplex containing substrates on P. aeruginosa RNase T. (A) Docking of an ideal A form RNA duplex with a two nucleotide 3′ overhang. While the unpaired 3′ overhang is shown stacked here, this region is likely to be distorted for hydrolytic cleavage. (B) Docking of an ideal B form DNA helix with a single nucleotide 3′ overhang. RNase T is shown as a surface potential diagram colored as in Figure 2, and substrates are shown as green ribbons. Only one substrate molecule was docked; the active pocket/RNA binding surface on the opposite side of RNase T is shown empty, and could presumably accommodate another substrate molecule in an analogous manner. Clashes in the docking models are mostly due to the long side chains of a few solvent exposed basic residues (Arg101, Arg114, Lys139 and Arg140). These sidechains are likely to adopt alternative conformations upon substrate binding.
