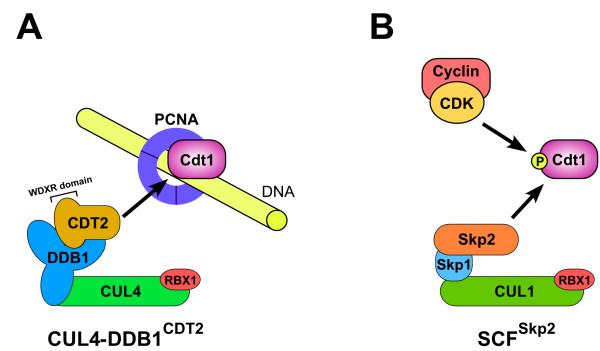
Figure 1.
Two distinct molecular pathways for Cdt1 degradation. (A and B) CUL4-DDB1CDT2 and SCFSkp2 CRL ubiquitin ligase complexes have similar modular structures: a cullin; a common RING H2-finger protein Rbx1; an adaptor protein, DDB1 or Skp1; and an SRS, CDT2 or Skp2. In the CUL4-DDB1CDT2 complex, CDT2 binds to DDB1 through a WD-repeat region with a specific signature, termed a 'WDXR' or 'DXR' domain (marked in figure). In the SCFSkp2 complex, Skp2 binds to Skp1 through an F-box motif (not marked). (A) CUL4-DDB1CDT2 targets Cdt1 for degradation after Cdt1 binds to PCNA on chromatin. CDT2 is proposed to directly bind Cdt1 after Cdt1 binds to PCNA, although the CDT2-Cdt1 interaction has not yet been formally demonstrated. (B) SCFSkp2 targets Cdt1 for degradation after CDK/Cyclin complexes phosphorylate Cdt1. See text for details.