Abstract
Azoarcus anaerobius, a strictly anaerobic, gram-negative bacterium, utilizes resorcinol as a sole carbon and energy source with nitrate as an electron acceptor. Previously, we showed that resorcinol degradation by this bacterium is initiated by two oxidative steps, both catalyzed by membrane-associated enzymes that lead to the formation of hydroxyhydroquinone (HHQ; 1,2,4-benzenetriol) and 2-hydroxy-1,4-benzoquinone (HBQ). This study presents evidence for the further degradation of HBQ in cell extracts to form acetic and malic acids. To identify the A. anaerobius genes required for anaerobic resorcinol catabolism, a cosmid library with genomic DNA was constructed and transformed into the phylogenetically related species Thauera aromatica, which cannot grow with resorcinol. By heterologous complementation, a transconjugant was identified that gained the ability to metabolize resorcinol. Its cosmid, designated R+, carries a 29.88-kb chromosomal DNA fragment containing 22 putative genes. In cell extracts of T. aromatica transconjugants, resorcinol was degraded to HHQ, HBQ, and acetate, suggesting that cosmid R+ carried all of the genes necessary for resorcinol degradation. On the basis of the physiological characterization of T. aromatica transconjugants carrying transposon insertions in different genes of cosmid R+, eight open reading frames were found to be essential for resorcinol mineralization. Resorcinol hydroxylase-encoding genes were assigned on the basis of sequence analysis and enzyme assays with two mutants. Putative genes for hydroxyhydroquinone dehydrogenase and enzymes involved in ring fission have also been proposed. This work provides the first example of the identification of genes involved in the anaerobic degradation of aromatic compounds by heterologous expression of a cosmid library in a phylogenetically related organism.
Resorcinol (1,3-dihydroxybenzene) is produced and utilized in large amounts by the timber, adhesives, and oil industries and enters freshwater environments through the release of effluents. Also, roots of aquatic plants such as Nuphar lutea exude resorcinol in considerable amounts into the aquatic environment (42). Resorcinol is photochemically transformed upon exposure to sunlight. However, these chemical modifications are, in general, slow processes (45) and do not degrade resorcinol to carbon dioxide. Therefore, most of the detoxification and reintroduction of resorcinol constituents back into the carbon cycle must be catalyzed by microbial degradation.
There have been several reports of bacteria able to utilize resorcinol as carbon and energy sources in the presence or absence of oxygen by different biochemical strategies. While aerobes dearomatize the benzene ring by using molecular oxygen as a cosubstrate for monooxygenases and dioxygenases, anaerobes have to replace all of the oxygen-dependent steps with alternative sets of reactions (6, 16, 21, 36). Notably, in anaerobic pathways, the aromatic ring is reduced rather than oxidized and benzoyl coenzyme A (CoA) emerges as the most common intermediate in the degradation of a large diversity of aromatic substrates (4, 5, 7, 21, 36, 46).
Since the late 1960s, several microorganisms such as Pseudomonas putida strain ORC (10), Trichosporon cutaneum (12), and Azotobacter vinelandii (18) were reported to be able to degrade resorcinol aerobically. In all of these cases, resorcinol is oxidized in reactions catalyzed by oxygenases to hydroxyhydroquinone (HHQ; 1,2,4-benzenetriol) or pyrogallol (1,2,3-benzenetriol). HHQ undergoes further diol ring cleavage with subsequent formation of maleylacetate and later β-ketoadipate (10, 12), while pyrogallol is cleaved to form oxalocrotonate and later pyruvate plus acetate (18). Very recently, a new Rhodococcus opacus strain named RW was isolated and shown to degrade resorcinol (29). So far, nothing is known about the biochemistry that R. opacus RW uses in the mineralization of resorcinol.
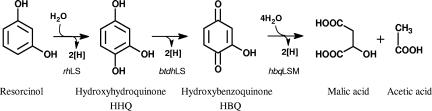
Anaerobic resorcinol degradation has been documented for various microorganisms, such as sulfate-reducing bacteria (38), fermenting bacteria (44), and denitrifiers (17, 26, 40). Notably, none of these bacteria use the benzoyl-CoA pathway in resorcinol catabolism. This is due to the fact that resorcinol as a dihydroxybenzene is less stabilized by resonance of the benzene ring and can be attacked reductively or oxidatively without prior activation. Both strategies have been described for resorcinol destabilization under anoxic conditions. While a fermenting Clostridium sp. uses reductive biochemistry to convert resorcinol to cyclohexanedione (26, 44), which is further hydrolyzed to 5-oxohexanoate (36), Anaerobius anaerobius, a nitrate-reducing bacterium, uses two oxidative reactions (32). Resorcinol is hydroxylated at position 4 of the aromatic ring with K3Fe(CN)6 or nitrate as an electron acceptor to form hydroxyhydroquinone (HHQ) in a reaction catalyzed by resorcinol hydroxylase. HHQ is subsequently oxidized with nitrate to HBQ (2-hydroxy-1,4-benzoquinone) in a reaction catalyzed by hydroxyhydroquinone dehydrogenase (32). Both reactions were measured in the membrane fraction of resorcinol-grown cells (32). The further fate of HBQ has not been studied yet. Hydroxylation of resorcinol in A. anaerobius provides a less energy-consuming alternative to the benzoyl-CoA pathway, in which benzoyl-CoA formation requires stoichiometric hydrolysis of ATP (46). Moreover, the electrons from resorcinol and HHQ oxidation can directly enter the denitrification process and may allow energy conservation by proton translocation (32).
The aims of our study were to further elucidate the resorcinol degradation pathway in A. anaerobius and to identify genes that code for resorcinol metabolism. Analysis of the sequences of 29.88-kb chromosomal DNA from a cosmid library should provide new insight into the function of the resorcinol-degrading enzymes.
MATERIALS AND METHODS
Chemicals.
All of the chemicals and biochemicals used in this study were purchased from Fluka (Neu Ulm, Germany), Merck (Darmstadt, Germany), Serva (Heidelberg, Germany), Sigma-Aldrich (Deisenhofen, Germany), or Biozym (Oldendorf, Germany) and were of the highest quality available. Enzymes used for cloning and other materials used for molecular biology techniques were obtained from MBI Fermentas (St. Leon-Rot, Germany), Roche Diagnostics (Mannheim, Germany), Eppendorf (Hamburg, Germany), Stratagene (Heidelberg, Germany), or QIAGEN (Hilden, Germany). 2-Hydroxy-1,4-benzoquinone (HBQ) was prepared nonenzymatically by auto-oxidation of hydroxyhydroquinone (10). Custom sequencing was performed at GATC (Constance, Germany).
Bacterial strains, vectors, and culture conditions.
The bacterial strains and vectors used in this work are summarized in Table 1. Azoarcus anaerobius strain LuFRes1, T. aromatica strains AR1 and K172, and their transconjugants were cultured anaerobically at 30°C in 100-ml infusion bottles containing 50 ml of nonreduced mineral medium (32) under nitrogen gas. The medium was buffered with 30 mM 3-(N-morpholino)propanesulfonic acid instead of bicarbonate and supplemented with 8 mM nitrate. Carbon sources were stored as 0.5 M stock solutions in sterile infusion bottles under nitrogen gas and added to cultures with syringes to final concentrations of 2 mM resorcinol (A. anaerobius), 2 mM 3,5-dihydroxybenzoate, 2 mM benzoate (T. aromatica AR1), and 5 mM succinate (T. aromatica K172). For biochemical studies, all strains were cultured in 1-liter infusion bottles and harvested at an optical density at 578 nm (OD578) of 0.3 to 0.4. To study resorcinol degradation and nitrate reduction by T. aromatica transconjugants containing pLAFR3 recombinant cosmid or EZ::TN<KAN-2> transposon insertional derivatives of cosmid R+, 100-ml serum bottles containing 50 ml of anaerobic mineral medium supplemented with resorcinol (2 mM) and nitrate (8 mM) were inoculated with 0.5% preculture. Escherichia coli HB101(pRK600) was used as a helper strain, and strains HB101 and DH5α (pLAFR3 derivative cosmids) were used as the donor strains in triparental matings. All E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (35). Antibiotics were used at the following concentrations: tetracycline, 20 μg ml−1; chloramphenicol, 30 μg ml−1; kanamycin, 50 μg ml−1.
TABLE 1.
Bacterial strains and cosmids used in this study
| Strain or vectors | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| A. anaerobius LuFRes1 (DSM12081) | Wild type; Res+ | 40 |
| T. aromatica | ||
| AR1 (DSM11528) | Wild type; Res− DHB+ HHQ pathway+ | 13 |
| K172 (DSM6984T) | Wild type; Res− DHB− HHQ pathway− | 2, 43 |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | 35 |
| HB101 | supE44 hsdS20(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | 35 |
| Plasmid pRK600 | ColE1 Tra+ Cmr | 22 |
| Cosmids | ||
| pLAFR3 | Tcr Tra− Mob+cos; RK2 replicon | 41 |
| R+ | Tcr, pLAFR3 containing a 29.9-kb resorcinol gene cluster of A. anaerobius | This study |
| Mu_4 | Tcr Kmr R+orf14::TN<KAN-2>; knockout mutant | This study |
| Mu_13 | Tcr Kmr R+btdhS::TN<KAN-2>; knockout mutant | This study |
| Mu_14 | Tcr Kmr R+orf12::TN<KAN-2>; knockout mutant | This study |
| Mu_16 | Tcr Kmr R+orf10::TN<KAN-2>; knockout mutant | This study |
| Mu_21 | Tcr Kmr R+rhL::TN<KAN-2>; knockout mutant | This study |
| Mu_22 | Tcr Kmr R+bqdhS::TN<KAN-2>; knockout mutant | This study |
| Mu_31 | Tcr Kmr R+orf3::TN<KAN-2>; knockout mutant | This study |
| Mu_36 | Tcr Kmr R+orf14::TN<KAN-2>; knockout mutant | This study |
| Mu_40 | Tcr Kmr R+rhS::TN<KAN-2>; knockout mutant | This study |
| Mu_41 | Tcr Kmr R+orf5::TN<KAN-2>; knockout mutant | This study |
| Mu_42 | Tcr Kmr R+orf7::TN<KAN-2>; knockout mutant | This study |
| Mu_44 | Tcr Kmr R+rhL::TN<KAN-2>; knockout mutant | This study |
| Mu_45 | Tcr Kmr R+orf10::TN<KAN-2>; knockout mutant | This study |
| Mu_46 | Tcr Kmr R+orf11::TN<KAN-2>; knockout mutant | This study |
| Mu_51 | Tcr Kmr R+bqdhL::TN<KAN-2>; knockout mutant | This study |
| Mu_53 | Tcr Kmr R+bqdhS::TN<KAN-2>; knockout mutant | This study |
| Mu_54 | Tcr Kmr R+orf2::TN<KAN-2>; knockout mutant | This study |
| Mu_56 | Tcr Kmr R+orf1::TN<KAN-2>; knockout mutant | This study |
| Mu_63 | Tcr Kmr R+orf9::TN<KAN-2>; knockout mutant | This study |
| Mu_64 | Tcr Kmr R+orf2::TN<KAN-2>; knockout mutant | This study |
| Mu_68 | Tcr Kmr R+orf2::TN<KAN-2>; knockout mutant | This study |
| Mu_73 | Tcr Kmr R+rhL::TN<KAN-2>; knockout mutant | This study |
| Mu_76 | Tcr Kmr R+btdhL::TN<KAN-2>; knockout mutant | This study |
| Mu_80 | Tcr Kmr R+bqdhL::TN<KAN-2>; knockout mutant | This study |
| Mu_82 | Tcr Kmr R+orf1::TN<KAN-2>; knockout mutant | This study |
| Mu_93 | Tcr Kmr R+orf13::TN<KAN-2>; knockout mutant | This study |
Res+, growth on resorcinol; Res−, no growth on resorcinol; DHB+, growth on 3,5-dihydroxybenzoate; DHB−, no growth on 3,5-dihydroxybenzoate; HHQ pathway+, has HHQ pathway; HHQ pathway−, lacks HHQ pathway; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance.
Growth and substrate depletion analysis.
Samples of the culture were withdrawn anaerobically with a sterile plastic syringe flushed with N2 at the designated time points. Growth was monitored spectrophotometrically by measuring OD578 in a Hitachi U-100 spectrophotometer. For substrate depletion analysis, the cells from the culture samples were pelleted (for 5 min by microcentrifuge) and the supernatant was stored at −20°C until analyzed by high-pressure liquid chromatography (HPLC) for resorcinol and/or 3,5-dihydroxybenzoate degradation, nitrate consumption, and nitrite formation as described below.
Preparation of cell extracts.
All of the steps in the preparation of cell extracts were performed anaerobically. Cultures of A. anaerobius and T. aromatica transconjugants were harvested at an OD578 of 0.3 and washed once under anoxic conditions with 100 ml of 50 mM potassium phosphate buffer (pH 7.0) as described previously (28). Unless used immediately, cell pellets were frozen in liquid N2 and stored at −20°C. To prepare cell extracts, cells were suspended in 50 mM potassium phosphate buffer (pH 7.0), passed through a French press at 138 MPa, and then centrifuged at 20,000 × g for 20 min at 4°C to remove cell debris. The membranes were collected by centrifugation at 100,000 × g for 1 h at 4°C and resuspended in anoxic 50 mM potassium phosphate buffer (pH 7.0) (half the volume of the cytosolic fraction). Both fractions were either used immediately or frozen in liquid N2 and stored at −20°C.
Protein determination.
Protein content was quantified by the Bradford method (8) with bovine serum albumin as the standard.
Enzymatic tests.
Resorcinol hydroxylase and hydroxyhydroquinone dehydrogenase enzyme activities were measured at 30°C under anoxic conditions as previously described (32), except that potassium phosphate buffer (50 mM, pH 7.0) was used as the buffer in all of the assays. A standard reaction mixture used to assay resorcinol hydroxylase contained 0.2 to 0.3 mg of protein, 1 mM K3Fe(CN)6, and 1 mM resorcinol. A standard reaction mixture used to assay hydroxyhydroquinone dehydrogenase contained 0.2 to 0.3 mg of protein, 1 mM NaNO3, and 1 mM resorcinol. NAD(P)H-dependent degradation of resorcinol beyond HBQ was coupled to nitrate reduction and monitored by measuring the formation of acetate, malate, and succinate by ion-exchange HPLC as described below. A standard reaction (1 ml) was started by addition of 1 mM resorcinol to the assay mixture containing the membrane fraction (1 mg protein ml−1), supplemented or not with the cytosolic fraction (1 mg protein ml−1), and incubated at 30°C in 50 mM potassium phosphate (pH 7.0) with 4 to 8 mM nitrate and 1 mM NADH. At defined time points, samples (25 μl) were withdrawn, mixed with ice-cold potassium phosphate buffer (25 μl), and immediately analyzed for resorcinol degradation by HPLC. Additional samples (150 μl) were withdrawn anoxically from the assay mixture, and the enzymatic reaction was stopped by adding 50 μl of 1 M sulfuric acid. Denatured proteins were removed by centrifugation, and the sample was analyzed immediately by HPLC for organic acid formation or stored at −20°C.
HPLC analysis.
Resorcinol, HHQ, and HBQ were analyzed with a C18 reversed-phase column (Grom-Sil 120 ODS-5ST, 5 μm, 150 by 4.6 mm; Grom, Herrenberg, Germany) as described elsewhere (32). Resorcinol degradation in cultures by T. aromatica transconjugants and resorcinol hydroxylase enzyme activities were measured discontinuously by HPLC with detection at 278 and 206 nm, respectively. The mobile phase, comprising a mixture of 100 mM ammonium phosphate buffer (pH 2.6) and methanol, was used at a flow rate of 1 ml min−1. The solvent phase (5% [vol/vol] methanol) was initially held for 1 min, and then the concentration was increased to 45% over a period of 6 min and then lowered within 0.5 min to 5% and held for an additional 7 min. Acetic and succinic acids were separated on an ion-exchange column (Aminex HPX-87H; Bio-Rad, Munich, Germany) that was operated at a flow rate of 0.6 ml min−1 at 40°C with 5 mM sulfuric acid solution as the mobile phase. Acetic acid eluted after 15.7 min, succinic acid eluted after 13.0 min, and malic acid eluted after 10.5 min. All three acids were quantified against external standards. Nitrate and nitrite were determined with an A06 anion-exchange column (3 by 120 mm; Sykam, Freising, Germany). A 40 mM NaCl solution was used as the eluent at a flow rate of 1 ml min−1. Nitrate eluted after 5.6 min, and nitrite eluted after 3.1 min. Both compounds were detected at 210 nm and quantified against external standards.
DNA manipulation.
Standard methods were used for genomic DNA preparation, DNA digestion with restriction endonucleases, ligation, agarose gel electrophoresis, and transformation of E. coli (35). Cosmids were prepared for sequencing and in vitro transposon mutagenesis experiments with the QIAGEN Large construct kit (QIAGEN, Hilden, Germany).
Cosmid library construction.
Genomic DNA of A. anaerobius was isolated by an established protocol (35). Partial digestion of the DNA with PstI was carried out, and fragments of 20 and 30 kb were isolated and ligated with the PstI-digested and dephosphorylated pLAFR3 (41) vector. The resulting ligation products were then packaged into lambda phage heads with a Gigapack III packaging extract (Stratagene). The phage particles were transduced into E. coli HB101, and colonies were grown on LB agar containing tetracycline (27). The resulting colonies were harvested in liquid LB medium, grown overnight, and used as the donors in triparental mating as described below, by using T. aromatica AR1 as the acceptor.
Triparental mating.
T. aromatica strains AR1 and K172 were used as recipients and grown for 60 h anaerobically in minimal medium supplemented with 3,5-dihydroxybenzoate (2 mM) and succinate (5 mM), respectively. The following E. coli strains were grown in LB medium overnight: strain HB101 containing helper plasmid pRK600 with chloramphenicol and strain HB1O1 and DH5α donors containing pLAFR3 recombinant cosmids with tetracycline or EZ::TN<KAN-2> transposon insertional derivatives of cosmid R+ with kanamycin. All steps in the triparental mating were done aerobically as follows. A 0.5-ml volume of E. coli culture and 3 ml of the recipient culture in the exponential growth phase were centrifuged at 13,000 × g for 10 min at 10°C. Cell pellets were washed twice with 1 ml LB medium. The resulting cell pellets of all three types of cells were resuspended together in 30 μl LB and distributed on a sterile 47-mm-diameter, 0.22-μm-pore-size filter (Schleicher & Schuell, Dassel, Germany) that was placed on an LB plate and incubated overnight at 30°C. The filter was transferred into 1 ml of mineral medium in a sterile tube. Cells were washed off by vigorous vortexing and transferred to 100-ml serum bottles containing 50 ml anoxic mineral medium supplemented with resorcinol (2 mM) and nitrate (8 mM). The cultures were incubated at 30°C for 1 to 3 weeks in the dark without shaking. The ability of these transconjugants to grow on resorcinol was confirmed by substrate depletion analysis.
Isolation of a T. aromatica AR1 transconjugant clone that degrades resorcinol.
Heterologous expression of an A. anaerobius cosmid library was achieved in T. aromatica strain AR1 at 30°C under anoxic conditions with resorcinol (2 mM) and nitrate (8 mM) as substrates in the presence of tetracycline. After 2 weeks of incubation, 2% of the grown cells were transferred into fresh anoxic medium supplemented with resorcinol (2 mM) and nitrate (8 mM) and allowed to continue to grow. After 2 days, the medium was turbid, indicating that resorcinol degradation took place. In order to isolate single transconjugant colonies, agar shake dilutions were performed as described elsewhere (31). Single transconjugant colonies were analyzed for colony shape and motility, and 16S rRNA gene analysis confirmed that the transconjugant was indeed T. aromatica AR1. The cosmid was isolated, sequenced, and designated R+. Cosmid R+ was propagated in E. coli strain HB101 and used as the donor in the second triparental mating with T. aromatica K172 as the acceptor.
Construction, isolation, and sequencing of transposon insertion mutants.
Gene knockouts were created by the in vitro transposon insertion mutagenesis technique with an EZ::TN<KAN-2> insertion kit (Epicenter, Oldendorf, Germany) and cosmid R+ as the template according to the manufacturer's instructions. E. coli DH5α competent cells were electroporated with the resulting transposon insertion products with a GenePulser (Bio-Rad, Munich, Germany). Transposon insertion clones were selected on kanamycin-containing LB plates since the transposon confers resistance to this antibiotic. To identify clones having a single transposon insertion at different positions in the coding region of cosmid R+, the cosmids of about 100 of the 3,000 colonies obtained were isolated and analyzed by restriction digestion with a selected set of restriction enzymes. Those cosmids considered interesting were introduced into T. aromatica AR1 and K172 via triparental mating, and the resulting transconjugants were grown in mineral medium supplemented with kanamycin, resorcinol, and nitrate as described above. Samples were withdrawn anaerobically with N2-flushed, sterile plastic syringes at the designated time points and analyzed for growth and resorcinol degradation. The precise sites of EZ::TN<KAN-2> transposon insertion were determined by sequencing the derivative cosmid R+ with transposon-specific primers provided in the in vitro transposon mutagenesis kit (KAN-2 FP-1 and KAN-2 RP-1).
Sequence analysis.
Nucleotide and amino acid sequences were analyzed with tools provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and the ExPASy molecular biology server (http://www.expasy.ch/). Transmembrane helices in proteins were predicted with TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Nucleotide sequence accession number.
The nucleotide sequence of the coding region of cosmid R+ was deposited in the NCBI (GenBank) nucleotide sequence database under accession no. EF078692.
RESULTS AND DISCUSSION
HBQ degradation in cell suspension.
Various attempts were made to establish culture conditions for in vivo degradation of the resorcinol degradation intermediates HHQ and HBQ in A. anaerobius. No growth was observed with HHQ or HBQ prepared by auto-oxidation of HHQ at concentrations between 0.5 and 2 mM. No degradation and no product formation were observed in experiments with dense suspensions of intact cells or with cells permeabilized with cetyltrimethylammonium bromide. These results suggest two things. Either (i) A. anaerobius has no uptake system for such polar compounds that are readily degraded once formed in the cell, or (ii) neither intermediate is an inducer in the resorcinol degradation pathway. Nevertheless, A. anaerobius was able to take up and metabolize 5 mM acetate, succinate, or malate, each of which is a carboxylic acid formed during resorcinol degradation (see below).
Identification of HBQ degradation products in cell extracts of A. anaerobius.
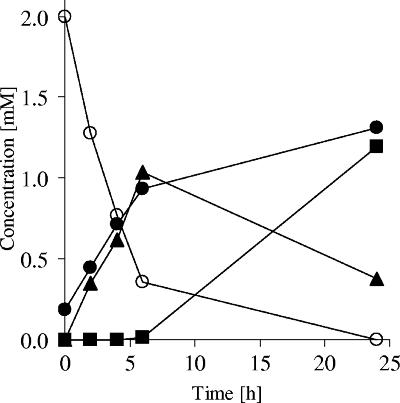
HBQ, the first nonaromatic intermediate in resorcinol degradation, is highly reactive (32, 47) and should be prone to ring fission. HBQ is not commercially available but can be produced by auto-oxidation of HHQ (10). In air, HBQ spontaneously oxidizes and forms polymers, which renders its detection by HPLC difficult. It also reacts with chemicals and components of cell extracts carrying thiol groups, giving rise to addition products. Therefore, HBQ depletion and product formation were studied with extracts prepared from resorcinol-grown cells which were incubated anoxically with resorcinol (1 to 2 mM) and nitrate (8 mM). In these experiments, nitrate was used as the electron acceptor for two reasons. In a previous study, it was shown that (i) oxidation of resorcinol and HHQ was dependent on electron acceptors with a positive redox potential such as K3Fe(CN)6 and nitrate (32) and (ii) nitrate was the only electron acceptor that did not react chemically with HHQ (32). Nitrate was used in excess to provide an electron acceptor for the first two reactions, namely, resorcinol hydroxylation and HHQ dehydrogenation, and to provide an electron acceptor for potential oxidations involved in the further degradation of HBQ. In assays with nitrate and membrane fractions prepared from resorcinol-grown cells, resorcinol was converted at an average rate of 2.1 mU mg−1. During the reaction, the assay mixture turned from colorless to brick red, which is indicative of HBQ accumulation (24, 30, 32), which was confirmed by HPLC analysis (32). No substrate degradation and no HBQ formation were detected in assay mixtures in which resorcinol or nitrate was omitted or in those containing membranes prepared from acetate-grown cells. These data are in agreement with the results previously reported (32). Addition of a cytosolic fraction of resorcinol-grown cells to assay mixtures containing membrane fractions of resorcinol-grown cells increased the rate of resorcinol degradation twofold (5.8 mU mg−1 protein). Nevertheless, HBQ accumulation was still revealed by red color development. Various coenzymes and cosubstrates, such as ascorbic acid, ATP, biotin, CoA, cysteine, dithiothreitol, glutathione, lipoic acid, naphthoquinone, thiamine pyrophosphate (TPP), NAD(P)+, NADPH, and NADH, were supplied in the assay mixtures containing resorcinol, nitrate, and both membrane and cytosolic fractions of resorcinol-induced cells. Except for NAD(P)H, none of them prevented HBQ accumulation. The resorcinol degradation rate increased to 9.9 mU mg−1 with either NADH or NADPH. HPLC chromatograms obtained by separation on an ion-exchange column displayed two products within the first 6 h of incubation, which increased proportionally with resorcinol consumption. One of the two metabolic products formed during the first 6-h reaction almost disappeared, and a third one appeared (Fig. 1). By comparison of their retention times with standards of different carboxylic acids and by coelution studies, the unknown metabolites were identified. The primary products were identified as acetic acid and malic acid, and the later one was identified as succinic acid. As shown in Fig. 1, almost stoichiometric amounts of succinic and acetic acids were formed from resorcinol. Acetate and malate were formed concomitantly as resorcinol was metabolized, and succinate was formed later while malate was consumed, indicating that malate was the primary reaction pr oduct. Control assay mixtures without resorcinol but with NADH showed that only negligible nitrate reduction took place. These results clearly indicated (i) that acetate, malate, and later succinate were formed in the resorcinol degradation pathway; (ii) that their formation was dependent on a soluble and resorcinol-induced enzyme(s) that required NAD(P)H; and (iii) that resorcinol could act as an inducer of all catabolic pathway genes (39). Similar findings were reported also for the anaerobic metabolism of other aromatic compounds by T. aromatica (9, 20, 37) and Azoarcus sp. strain EbN1 (34).
FIG. 1.
Utilization of resorcinol (○) and formation of succinate (▪), acetate (•), and malate (▴) by a mixture of the membrane fraction and cytosolic fraction of resorcinol-grown A. anaerobius.
Cosmid R+ contains all of the genes involved in resorcinol degradation by A. anaerobius.
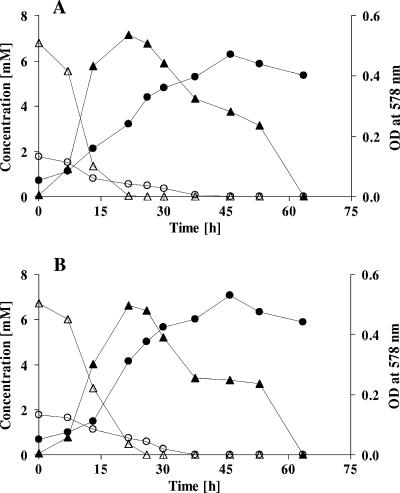
A. anaerobius and T. aromatica belong to the class Betaproteobacteria and are capable of growing at the expense of aromatic compounds under denitrifying conditions (32, 33, 40). A cosmid library with A. anaerobius chromosomal DNA was constructed and heterologously expressed in two T. aromatica strains, AR1 and K172, that are unable to metabolize resorcinol. Nevertheless, T. aromatica AR1 degrades α-resorcylate (3,5-dihydroxybenzoate) via the HHQ pathway (14, 33) and can therefore be expected to harbor genes similar to those of A. anaerobius. T. aromatica K172 is known to degrade neither resorcinol nor α-resorcylate. Transconjugants of both strains containing cosmid R+ were able to grow with resorcinol even after seven successive passages without resorcinol and the selective marker tetracycline (data not shown), demonstrating that cosmid R+ was stable in both hosts. T. aromatica transconjugants degraded resorcinol at a rate similar to that of A. anaerobius (about 1 mM resorcinol day−1) (data not shown). The increase in OD578 was coupled to resorcinol degradation and to a simultaneous reduction of nitrate to nitrite by both transconjugants (Fig. 2A and B), indicating that at least the resorcinol hydroxylase gene(s) should be contained in cosmid R+, and this was confirmed by enzymatic tests. Resorcinol hydroxylase was measured in membrane fractions of both transconjugants prepared from cells grown with resorcinol but not from cells grown with an alternative substrate (Table 2). These results are in good agreement with previously reported data (32). As expected, no resorcinol hydroxylase was found in either wild-type strain of T. aromatica. To determine if the HHQ dehydrogenase-encoding gene(s) is on the cosmid R+, enzyme activities were measured in transconjugants and compared with those found in the wild-type strains. HHQ dehydrogenase activity was not detected in T. aromatica K172 (wild type), as expected, but was found in all other cell extracts (nonfractionated by ultracentrifugation) of both transconjugants grown on resorcinol or on a different aromatic substrate. In both transconjugants, the activities measured were in the same range as those measured in A. anaerobius. A twofold higher HHQ dehydrogenase activity than in A. anaerobius extracts was detected in cell extracts prepared from an AR1 transconjugant grown with α-resorcylate. Although A. anaerobius is unable to grow with α-resorcylate, this finding can be explained only by its structural similarity to resorcinol, which may make it act as a good gratuitous inducer of HHQ dehydrogenase. In this context, benzoate, which carries no hydroxyl groups, may act as a bad gratuitous inducer. It will be interesting to determine if other dihydroxybenzoates, trihydroxybenzenes, and trihydroxybenzoate isomers can act as inducers of both HHQ dehydrogenases of A. anaerobius and T. aromatica AR1. To determine if also the ring cleavage enzyme(s) is encoded by cosmid R+, membrane fractions of transconjugants grown with resorcinol were mixed with cytosolic fractions of the respective transconjugants grown with either resorcinol or an alternative carbon source and tested for acetate, malate, and succinate formation. These assay mixtures contained HHQ (1 mM), nitrate (4 mM), and NADH (1 mM). Substoichiometri c amounts of acetate were formed in all assay mixtures containing cytosolic fractions of resorcinol-grown transconjugants. After 6 h, 0.21 mM acetate was formed with the cytosolic fraction of resorcinol-grown cells of strain AR1/R+ and 0.11 mM acetate was formed with the cytosolic fraction of strain K1721/R+. No malate or succinate was detected in these assay mixtures. No acetate was formed in the assay mixtures containing cytosolic fractions prepared from cells grown without resorcinol. These results demonstrate that the genes for the whole resorcinol degradation pathway are encoded by cosmid R+.
FIG. 2.
Degradation of resorcinol by transconjugants T. aromatica AR1 (A) and K172 (B) harboring cosmid R+. Resorcinol (○) is metabolized with concomitant consumption of nitrate (▵), release of nitrite (▴), and an increase in OD578 (•).
TABLE 2.
Specific activities of resorcinol hydroxylase in membrane fractions and HHQ dehydrogenase in unfractionated cell extracts of A. anaerobius and T. aromatica strains
| Strain | Growth substrate | Sp act (mU mg−1 protein)
|
|
|---|---|---|---|
| Resorcinol hydroxylase | HHQ dehydrogenase | ||
| A. anaerobius | Resorcinol | 98 | 33 |
| Benzoate | 0 | 8 | |
| T. aromatica AR1/R+ | Resorcinol | 80 | 37 |
| α-Resorcylate | 0 | 80 | |
| T. aromatica K172/R+ | Resorcinol | 150 | 20 |
| Benzoate | 0 | 3 | |
| T. aromatica AR1 wild type | α-Resorcylate | 0 | 27 |
| T. aromatica K172 wild type | Benzoate | 0 | 0 |
Genes and gene organization on cosmid R+.
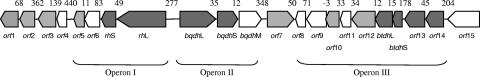
Sequencing of both strands of the coding region of cosmid R+ resulted in sequence information for 29.88 kb, which comprised 22 open reading frames (ORFs). Table 3 and Figure 3 summarize the locations of all 22 genes, the number of amino acids encoded by each protein, and the percent similarity and percent identity of the amino acid sequences of all 22 proteins with known sequences in the databases. No stop codon was detected for orf1, indicating that this gene was incomplete. Except for btdhL, all of the other ORFs were preceded by putative, typical Shine-Dalgarno ribosomal binding sites, which are very similar to the consensus sequence AGGAGG located 4 to 13 nucleotides upstream of the starting codon. On the basis of a BLAST similarity search, an alternative start codon, GTG, was identified for orf3. Except for orf3, in all of the genes translation is initiated with ATG. Taking into account the ORF's direction of transcription and the size of the intergenic spaces, it appears that the coding region of cosmid R+ is organized into three putative operons that may form three transcriptional units. In the products encoded by orf2, orf9, orf11, and btdhS, 1, 12, 2, and 4 transmembrane helices were predicted, respectively. The analysis of the 29.88-kb coding region of cosmid R+ allowed us to tentatively assign functions to 20 out of 22 gene products—except that it showed that cosmid R+ contains all of the elements required for high-affinity nutrient acquisition systems (three transporters), detoxification (one glutathione-S-transferase), catabolism (10 oxidoreductases), and components with regulatory functions (σ factors, regulators, and a stress response system).
TABLE 3.
Properties of genes and gene products encoded by cosmid R+
| Gene product | Gene properties
|
Protein properties
|
Protein correspondence
|
Protein with highest similarity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Range | % GC | Length (amino acids) | Mol mass (kDa) | pI | % Identity | % Similarity | E value | Reference | ||
| ORF1 | 1-1150 | 68 | 384 | 41.8 | 8.9 | 59 | 70 | 9e-104 | gi 121608751 ref YP_996558.1 | Transposase (Verminephrobacter eiseniae) |
| ORF2 | 1218-2147 | 63 | 309 | 33.3 | 6.0 | 61 | 77 | 1e-94 | gi 118592793 ref ZP_01550182.1 | ABC transporter substrate-binding protein (Stappia aggregata) |
| ORF3 | 2509-3618 | 67 | 369 | 38.9 | 6.3 | 85 | 90 | 1e-95 | gi 56479427 ref YP_161016.1 | Possibly flavin-dependent dehydrogenase (Azoarcus sp. strain EbN1) |
| ORF4 | 3757-4656 | 57 | 299 | 32.9 | 6.2 | 72 | 84 | 8e-121 | gi 56478704 ref YP_160293.1 | Possibly involved in regulation of phenolic degradation (Azoarcus sp. strain EbN1) |
| ORF5 | 5096-5701 | 62 | 201 | 22.7 | 5.3 | 70 | 78 | 1e-68 | gi 89358401 ref ZP_01196223.1 | Conserved hypothetical protein (Xanthobacter autotrophicus) |
| ORF6 | 5712-6680 | 62 | 322 | 36.7 | 6.7 | 61 | 72 | 1e-109 | gi 110634534 ref YP_674742.1 | Putative glutathione S-transferase (Mesorhizobium sp.) |
| RHS | 6763-7659 | 62 | 298 | 33.2 | 7.5 | 55 | 70 | 5e-92 | gi 118698587 ref ZP_01556663.1 | Pyrogallol hydroxytransferase (Burkholderia ambifaria) |
| RHL | 7708-10455 | 63 | 915 | 102.7 | 6.6 | 67 | 66 | 0.0 | gi 118698588 ref ZP_01556664.1 | Pyrogallol hydroxytransferase (Burkholderia ambifaria) |
| BQDHL | 11332-13554 | 65 | 740 | 81.1 | 5.6 | 57 | 73 | 0.0 | gi 119946408 ref YP_944088.1 | Pyruvate dehydrogenase (Psychromonas ingrahamii) |
| BQDHS | 13589-14854 | 69 | 421 | 43.3 | 5.9 | 55 | 70 | 2e-102 | gi 83593214 ref YP_426966.1 | Dihydrolipoamide acetyltransferase (Rhodospirillum rubrum) |
| BQDHM | 14866-16263 | 65 | 465 | 49.6 | 6.2 | 68 | 83 | 3e-180 | gi 23013388 ref ZP_00053288.1 | Pyruvate/2-oxoglutarate dehydrogenase (Magnetospirillum magnetotacticum) |
| ORF7 | 16611-18260 | 68 | 549 | 60.3 | 7.7 | 38 | 59 | 6e-110 | gi 56419244 ref YP_146562.1 | Acetoin operon expression regulatory protein (Geobacillus kaustophilus) |
| ORF8 | 18310-18933 | 61 | 207 | 23.8 | 5.4 | 38 | 53 | 2e-38 | gi 88940881 ref ZP_01146311.1 | Putative protein-disulfide isomerase (Acidiphilium cryptum) |
| ORF9 | 19004-20239 | 65 | 411 | 42.6 | 9.8 | 54 | 69 | 3e-76 | gi 71907514 ref YP_285101.1 | Major facilitator superfamily MFS_1 (Dechloromonas aromatica) |
| ORF10 | 20236-21306 | 63 | 356 | 38.8 | 5.5 | 44 | 58 | 2e-66 | gi 83308699 emb CAJ01609.1 | Cobalamin synthesis protein cobW/p47k family protein (Methylocapsa acidiphila) |
| ORF11 | 21339-21920 | 65 | 193 | 21.8 | 7.1 | 54 | 67 | 2e-38 | gi 91790635 ref YP_551587.1 | Antibiotic biosynthesis monooxygenase (Polaromonas sp.) |
| ORF12 | 21954-23414 | 65 | 486 | 51.9 | 5.6 | 73 | 86 | 0.0 | gi 118744335 ref ZP_01592329.1 | Succinic semialdehyde dehydrogenase (Geobacter lovleyi) |
| BTDHL | 23426-24336 | 68 | 303 | 31.3 | 6.0 | 45 | 63 | 4e-46 | gi 27377791 ref NP_769320.1 | Dehydrogenase (Bradyrhizobium japonicum) |
| BTDHS | 24352-24957 | 60 | 201 | 22.2 | 10.4 | 39 | 53 | 1e-14 | gi 78693872 ref ZP_00858386.1 | Hypothetical protein (Bradyrhizobium sp.) |
| ORF13 | 25135-26388 | 65 | 417 | 46.6 | 5.5 | 48 | 68 | 1e-104 | gi 119946410 ref YP_944090.1 | Peptidase M24 (Psychromonas ingrahamii) |
| ORF14 | 26433-27548 | 61 | 371 | 40.8 | 6.4 | 51 | 67 | 3e-102 | gi 74317595 ref YP_315335.1 | NADH-dependent flavin oxidoreductase (Thiobacillus denitrificans) |
| ORF15 | 27752-29737 | 68 | 661 | 72.8 | 6.9 | 37 | 57 | 3e-117 | gi 114567214 ref YP_754368.1 | Acetoin operon expression regulatory protein (Syntrophomonas wolfei) |
FIG. 3.
Organization of the 29,880-bp gene cluster of cosmid R+ involved in anaerobic resorcinol metabolism in A. anaerobius. Arrows indicate the direction of translation of each ORF. Genes encoding enzymes required for resorcinol degradation are shaded dark gray. Genes shown to be nonessential for resorcinol degradation in growth experiments with transposon insertion mutants are in light gray, and the genes encoding proteins so far unknown to be essential or nonessential for resorcinol degradation are unshaded. Sizes of gaps between genes are indicated in base pairs above the schematically drawn ORFs.
Isolation of mutants deficient in resorcinol utilization.
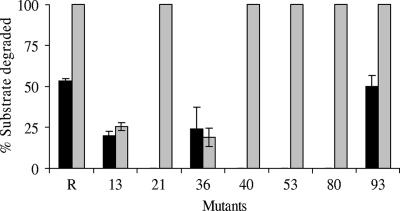
To analyze the resorcinol degradation pathway of A. anaerobius in greater detail, in vitro transposon mutagenesis was carried out with cosmid R+ by using the EZ::TN<KAN2> transposon. This particular transposon is a very useful tool for studies aiming to identify essential genes since no polar effects of transposon insertions on downstream genes can take place. This is due to the fact that the transposon contains multiple E. coli-type promoters oriented outward in both directions (15). This procedure generated a reasonable number (3,000) of stable mutated cosmids in E. coli, 100 of which were isolated and sequenced. Sequencing of the transposon insertion sites revealed a duplication of 9 bp at each point of insertion, which is consistent with the properties of the EZ::TN<KAN2> transposon. Only 26 out of the 100 transposon insertional cosmids initially selected were found to contain one transposon inserted in the A. anaerobius chromosomal DNA of cosmid R+. Both strains of T. aromatica, AR1 and K172, were transformed by mating with the selected 26 cosmid R+ derivatives and screened for the ability to grow on resorcinol. In repeated experiments with at least three independent matings, we confirmed that 12 mutants, T. aromatica AR1 and K172 transconjugants, were unable to grow with resorcinol (2 mM) but were able to grow in the presence of kanamycin on benzoate (2 mM) and succinate (5 mM), respectively. These results strongly indicate that the failure to metabolize resorcinol was not due to an unsuccessful transconjugation but rather was caused by the disruption of key enzymes involved in resorcinol degradation. Interestingly, cultures of AR1 transconjugants containing Mu_13 and Mu_36 cosmids developed a red-orange color during incubation with resorcinol which indicated that HHQ or HBQ was formed. In addition to the 12 mutants of T. aromatica K172, 1 more mutant, Mu_76, was also impaired in resorcinol degradation and growth with resorcinol. Therefore, we assume that this mutant had a defect in the gene encoding HHQ dehydrogenase, which in strain AR1 could be complemented by its chromosomal gene. Since it was shown in this study that α-resorcylate could induce A. anaerobius HHQ dehydrogenase from cosmid R+, we believe that resorcinol can act as an inducer of the T. aromatica AR1 chromosomal gene. Table 4 summarizes the transposon insertion sites in the coding region of cosmid R+ and the phenotypes of T. aromatica AR1 and K172 transconjugants carrying mutated cosmids. To determine if host chromosomal genes could complement any of the defects carried by the 12 T. aromatica AR1 mutants unable to grow on resorcinol, we monitored the resorcinol consumption of mutants grown with nitrate (8 mM) on mixtures of resorcinol (2 mM) and α-resorcylate (2 mM) over a 1-week period. These experiments were carried out with only one mutant for each key gene that had the transposon inserted nearest to the N terminus of the gene. In Fig. 4, the results obtained at the end of 4 days of growth are shown since after that the medium was almost depleted of the electron acceptor. The mutants were grouped into three classes. Mu_93 was able to metabolize both substrates at the same time but at different rates (data not shown). The Mu_21, Mu_40, Mu_53, and Mu_80 mutants did not degrade resorcinol at all but did degrade α-resorcylate. The Mu_13 and Mu_36 mutants grew poorly and degraded both substrates partially at the same time. In addition, these cultures accumulated a red-orange color which was observed as well in the cultures incubated with resorcinol only. However, no intermediate was detected by HPLC, which might mean that either it was not stable and decayed during sample preparation, analysis, or storage or that it was present in undetectable amounts in the culture samples analyzed. Studies concerning these two mutants are in progress.
TABLE 4.
Transposon insertion sites in the genes of cosmid R+ and mutant phenotypes in recombinant T. aromatica strains AR-1 and K172
| Mutant | Transposon insertion site | Growtha of strain K172
|
Growtha of strain AR1
|
||
|---|---|---|---|---|---|
| Resorcinol (2 mM) | Succinate (2 mM) | Resorcinol (2 mM) | Benzoate (2 mM) | ||
| Mu_4 | 27028 | − | + | − | + |
| Mu_13 | 24529 | − | + | − | + |
| Mu_14 | 22541 | + | ND | + | ND |
| Mu_16 | 21134 | + | + | + | ND |
| Mu_21 | 10148 | − | + | − | + |
| Mu_22 | 14692 | − | + | − | + |
| Mu_31 | 3212 | + | ND | + | ND |
| Mu_36 | 27319 | − | + | − | + |
| Mu_40 | 7196 | − | + | − | + |
| Mu_41 | 5656 | + | ND | + | ND |
| Mu_42 | 17257 | + | ND | + | ND |
| Mu_44 | 9933 | − | + | − | + |
| Mu_45 | 21277 | + | ND | + | ND |
| Mu_46 | 21371 | + | ND | + | ND |
| Mu_51 | 12756 | − | + | − | + |
| Mu_53 | 13965 | − | + | − | + |
| Mu_54 | 1438 | + | ND | + | ND |
| Mu_56 | 924 | + | ND | + | ND |
| Mu_63 | 19259 | + | ND | + | ND |
| Mu_64 | 1466 | + | ND | + | ND |
| Mu_68 | 1527 | + | ND | + | ND |
| Mu_73 | 8306 | − | + | − | + |
| Mu_76 | 24230 | − | + | + | ND |
| Mu_80 | 11364 | − | + | − | + |
| Mu_82 | 925 | + | ND | + | ND |
| Mu_93 | 26058 | − | + | − | + |
+, growth; −, no growth; ND, not determined.
FIG. 4.
Percentage of carbon sources utilized by T. aromatica AR1 mutants at the end of 4 days of incubation with a mixture of resorcinol (black bars)and α-resorcylate (gray bars).
Functional assignment of the identified key gene products in resorcinol metabolism.
In the present study, we have obtained biochemical evidence of NAD(P)H- and nitrate-dependent reactions involved in further degradation of HBQ to acetate, malate, and succinate. HHQ, the precursor of HBQ, is a common intermediate in the aerobic degradation of various aromatic compounds, such as resorcinol by T. cutaneum (12), P. putida (10), or Corynebacterium glutamicum (23); 4-nitrophenol by Arthrobacter sp. (24); 4-nitrocatechol by Burkholderia cepacia (11); and chlorohydroxyquinol by B. cepacia AC1100 (47). For all of the above pathways, except 4-nitrocatechol, it has been shown or proposed that the hydroxyhydroquinone intermediate is converted to maleylacetate, which is subsequently transformed to β-ketoadipate, which is cleaved to succinate and acetate, which enter the trichloroacetic acid cycle. In the aerobic degradation of 4-nitrocatechol, hydroxyhydroquinone is converted to 1,4-benzenediol (hydroquinol) and γ-hydroxymuconic semialdehyde. Maleylacetate reductase and CoA transferase, two enzymes characteristic of aerobic metabolism, were found to be insignificantly low and unstable in A. anaerobius (32). Moreover, none of the proteins encoded by cosmid R+ showed similarity to the corresponding proteins involved in the aerobic degradation of resorcinol or HHQ. All of these findings strongly indicate that A. anaerobius uses for resorcinol degradation a set of enzymes that differ from that which aerobes use when dealing with either resorcinol or HHQ. Mapping of transposon insertions in cosmid R+ derivatives of those mutants impaired in resorcinol utilization revealed that eight ORFs code for enzymes specific for resorcinol degradation in A. anaerobius. These products are encoded by rhLS, bqdhLS, btdhLS, orf13, and orf14.
Genes of resorcinol hydroxylase.
The product of rhLS was identified as resorcinol hydroxylase by monitoring the growth and resorcinol degradation of T. aromatica transconjugants containing the cosmid R+ derivates Mu_21, Mu_40, Mu_ 44, and Mu_73. In addition, mutants Mu_21 and Mu_40 were grown on resorcinol and α-resorcylate and the membrane fractions were prepared and confirmed for resorcinol hydroxylase activity. The membrane fraction of a transconjugant containing cosmid R+ that was grown and prepared in the same way was used as a positive control. Neither activity nor color development was detected in assay mixtures with membranes prepared from mutants grown with resorcinol and α-resorcylate (Table 5). According to BLAST analysis, the products of rhL and rhS are similar to anaerobic molybdopterin oxidoreductases and anaerobic dimethyl sulfoxide reductase-like enzymes. The best match for the rhL product to a biochemically studied enzyme was 52% identity to the α subunit of Pelobacter acidigallici pyrogallol-phloroglucinol transhydroxylase, and for the rhS product it was 49% identity to the β subunit of the same P. acidigallici transhydroxylase. Pyrogallol-phloroglucinol transhydroxylase is a soluble protein that catalyzes the conversion of pyrogallol to phloroglucinol (1,3,5-trihydroxybenzene). Its sequence is available (3), and its crystal structure was resolved as well (1, 28). The holoenzyme contains a molybdenum ion coordinated to two molybdopterin guanidine dinucleotide cofactors in the large subunit and three four-iron, four-sulfur clusters in the small subunit (3, 28). Binding sites for molybdenum and two molybdopterin guanidine dinucleotide molecules were identified in the rhL product, and eight conserved cysteine residues that could coordinate two iron-sulfur clusters were found in the rhS product. The cellular localization of this enzyme is still an unsolved problem. Resorcinol hydroxylase is predicted from the amino acid sequence to be soluble; however, it was measured almost exclusively in the membranes of A. anaerobius (32) and T. aromatica transconjugants. One may speculate that it could interact with an as-yet-unknown membrane anchor. On the basis of all of these findings, we propose that rhL and rhS indeed encode resorcinol hydroxylase. The resorcinol hydroxylase of A. anaerobius is the first anaerobic hydroxylase that converts resorcinol through hydroxylation and was characterized at the genetic level in this study.
TABLE 5.
Specific activities of resorcinol hydroxylase in the membrane fraction of T. aromatica AR1 transconjugants
| Strain | Growth substrate | Resorcinol hydroxylase sp act (mU mg−1 protein) |
|---|---|---|
| T. aromatica AR1/R+ | Resorcinol-α-resorcylate | 31 |
| T. aromatica AR1/Mu_21 | Resorcinol-α-resorcylate | 0 |
| T. aromatica AR1/Mu_40 | Resorcinol-α-resorcylate | 0 |
Genes of HHQ dehydrogenase.
Mutant T. aromatica K172 carrying the cosmid designated Mu_76 lost the ability to grow on resorcinol, while mutant T. aromatica AR1 carrying the same derivative of the R+ cosmid was not affected by this mutation at all. A reasonable explanation for these phenotypes was that the btdhL product was involved in the conversion of HHQ to HBQ. BLAST analysis of the product of btdhL showed that it is similar to putative β-hydroxy acid dehydrogenases of various genera belonging to proteobacteria, suggesting that all of these proteins may have similar mechanistic aspects. The missing ribosomal binding sites of btdhL could be part of a downregulation strategy for this gene, with the aim of securing just a low level of the corresponding protein. Upstream of btdhL, a small gene (btdhS) was identified which encodes a membrane protein of unknown function. Disruption of btdhS by a transposon insertion resulted in mutant Mu_13. Neither AR1 nor K172 transconjugants carrying the Mu_13 cosmid were able to grow on resorcinol alone. However, T. aromatica AR1 transconjugants accumulated a red-orange color that indicated negligible amounts of HHQ or HBQ formation. Since HHQ dehydrogenase was measured in the membrane fraction of A. anaerobius (32) and this study showed that both of the proteins encoded by btdhLS were required for resorcinol degradation, we conclude that btdhLS encodes the HHQ dehydrogenase.
Ring cleavage enzymes.
Disruption of bqdhL and bqdhS by a transposon insertion resulted in the generation of mutants Mu_22, Mu_51, Mu_53, and Mu_80. Neither AR1 nor K172 transconjugants carrying any of the above derivative cosmids were able to grow on resorcinol alone or to degrade resorcinol in cultures containing, in addition, α-resorcylate, indicating that both encode key enzymes in the resorcinol pathway and have no homologues in T. aromatica AR1. The bqdhL and bqdhS products aligned with proteins similar to the E1 and E2 components of multicomponent enzymes such as acetoin dehydrogenase, pyruvate dehydrogenase, and branched-chain α-keto acid dehydrogenase. E1 components of these enzymes require TPP and catalyze either an irreversible oxidative decarboxylation or a two-carbon unit transfer. Near the N terminus of the bqdhL product, a conserved TPP-binding motif (19) was identified. In the bqdhS product, no lipoyl-binding domain was found. A protein similar to the E3 component of pyruvate dehydrogenase or 2-oxoglutarate dehydrogenase is encoded by bqdhM, located downstream of bqdhS, for which no mutant is available yet. Two characteristic motifs for proteins likely to adopt a Rossmann fold and bind to a flavin adenine dinucleotide or NADP cofactor (25) were located near the N terminus of this protein. However, no experimental evidence for an involvement of a TPP, lipoyl, or flavin adenine dinucleotide cofactor in resorcinol degradation has been obtained so far. Nevertheless, on the basis of sequence similarity, we propose that the bqdhL, bqdhS, and bqdhM products are probably involved in the ring cleavage reaction. While the substrate of this enzyme is not known, malate and acetate could be the products of this reaction. In this study, it was shown that malate and acetate were formed simultaneously when resorcinol was metabolized. The conversion of HBQ to acetate and malate requires the splitting of two C-C bonds and releases two electrons. Such an oxidative cleavage reaction is compatible with an enzyme system analogous to a pyruvate dehydrogenase complex. The role of NAD(P)H in the ring cleavage reaction remains unknown.
Two genes with enigmatic functions are key enzymes in the resorcinol pathway.
The putative product of orf14 showed similarity to putative NADH-dependent flavin oxidoreductases of β- and α-proteobacteria such as Thiobacillus denitrificans and Bradyrhizobium japonicum. Trimethylamine dehydrogenase, 2,4-dienoyl-CoA reductase, enoate reductase, pentaerythritol tetranitrate reductase, xenobiotic reductase, and morphinone reductase are such oxidoreductases. It is suggested that these proteins contain a noncovalently bound flavin mononucleotide cofactor and iron sulfur clusters and that they use NAD(P)H as an electron donor. Disruption of orf14 by transposon insertion resulted in the generation of two mutated cosmids designated Mu_4 and Mu_36. Growth experiments with T. aromatica AR1 and K172 transconjugants containing these cosmids showed that its product is essential in resorcinol degradation and could be complemented to some extent by a T. aromatica AR1 protein. The function of this protein is not clear; it could be involved in electron transfer. A BLAST search revealed high similarities of the orf13 product with aminopeptidases, members of family M24, from Psychromonas ingrahamii and Acidothermus cellulolyticus (48% sequence identity). Such enzymes have been shown to be essential for removal of the initiating methionine of many proteins that are involved in protein maturation. If this is the case in A. anaerobius is not clear. If it is, we assume that it might be involved in the maturation of enzymes such as resorcinol hydroxylase or HHQ dehydrogenase. Our hypothesis is also based on the fact that possible chaperons were identified on cosmid R+.
To summarize the current status, we propose a tentative model showing a pathway correlated with the putative function of the key gene products (Fig. 5). Having at hand the sequences of all of the genes will allow us to heterologously produce A. anaerobius enzymes in E. coli to facilitate a detailed study of each protein and to finally elucidate the degradation pathway. Such studies are under way in our laboratory.
FIG. 5.
Proposed resorcinol pathway and putative functions of gene products.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany, and by research funds of the University of Constance. Work in Spain was financed by grant VEM2003-20075-CO2-01 from the Spanish Ministry of Science and Education. JIM-B work in Germany was partially supported by a Spanish-German exchange program (Acciones Integradas) of the Ministry of Science and Education.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Abt, D. J., O. Einsle, H. Niessen, R. Krieger, A. Messerschmidt, B. Schink, and P. M. Kroneck. 2002. Crystallization and preliminary X-ray analysis of the molybdenum-dependent pyrogallol-phloroglucinol transhydroxylase of Pelobacter acidigallici. Acta Crystallogr. D Biol. Crystallogr. 58:343-345. [DOI] [PubMed] [Google Scholar]
- 2.Anders, H.-J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 3.Baas, D., and J. Retey. 1999. Cloning, sequencing and heterologous expression of pyrogallol-phloroglucinol transhydroxylase from Pelobacter acidigallici. Eur. J. Biochem. 265:896-901. [DOI] [PubMed] [Google Scholar]
- 4.Boll, M. 2005. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10:132-142. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M. 2005. Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta 1707:34-50. [DOI] [PubMed] [Google Scholar]
- 6.Boll, M., and G. Fuchs. 2005. Unusual reactions involved in anaerobic metabolism of phenolic compounds. Biol. Chem. 386:989-997. [DOI] [PubMed] [Google Scholar]
- 7.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Breinig, S., E. Schiltz, and G. Fuchs. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J. Bacteriol. 182:5849-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauhan, A., S. K. Samanta, and R. K. Jain. 2000. Degradation of 4-nitrocatechol by Burkholderia cepacia: a plasmid-encoded novel pathway. J. Appl. Microbiol. 88:764-772. [DOI] [PubMed] [Google Scholar]
- 12.Gaal, A., and H. Y. Neujahr. 1979. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J. Bacteriol. 137:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallus, C., N. Gorny, W. Ludwig, and B. Schink. 1997. Anaerobic degradation of α-resorcylate by a nitrate-reducing bacterium, Thauera aromatica strain AR-1. Syst. Appl. Microbiol. 20:540-544. [Google Scholar]
- 14.Gallus, C., and B. Schink. 1998. Anaerobic degradation of α-resorcylate by Thauera aromatica strain AR-1 proceeds via oxidation and decarboxylation to hydroxyhydroquinone. Arch. Microbiol. 169:333-338. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 17.Gorny, N., G. Wahl, A. Brune, and B. Schink. 1992. A strictly anaerobic nitrate-reducing bacterium growing with resorcinol and other aromatic compounds. Arch. Microbiol. 158:48-53. [DOI] [PubMed] [Google Scholar]
- 18.Groseclose, E. E., and D. W. Ribbons. 1981. Metabolism of resorcinylic compounds by bacteria: new pathway for resorcinol catabolism in Azotobacter vinelandii. J. Bacteriol. 146:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins, C. F., A. Borges, and R. N. Perham. 1989. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 20.Heider, J., M. Boll, K. Breese, S. Breinig, C. Ebenau-Jehle, U. Feil, N. Gad'on, D. Laempe, B. Leuthner, M. E. Mohamed, S. Schneider, G. Burchhardt, and G. Fuchs. 1998. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170:120-131. [DOI] [PubMed] [Google Scholar]
- 21.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243:577-596. [DOI] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y., K. X. Zhao, X. H. Shen, M. T. Chaudhry, C. Y. Jiang, and S. J. Liu. 2006. Genetic characterization of the resorcinol catabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:7238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain, R. K., J. H. Dreisbach, and J. C. Spain. 1994. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 60:3030-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleiger, G., and D. Eisenberg. 2002. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through Cα-H…O hydrogen bonds and van der Waals interactions. J. Mol. Biol. 323:69-76. [DOI] [PubMed] [Google Scholar]
- 26.Kluge, C., A. Tschech, and G. Fuchs. 1990. Anaerobic metabolism of resorcyclic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch. Microbiol. 155:68-74. [Google Scholar]
- 27.Kretz, P. L., S. W. Kohler, and J. M. Short. 1991. Identification and characterization of a gene responsible for inhibiting propagation of methylated DNA sequences in mcrA mcrB1 Escherichia coli strains. J. Bacteriol. 173:4707-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messerschmidt, A., H. Niessen, D. Abt, O. Einsle, B. Schink, and P. M. Kroneck. 2004. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc. Natl. Acad. Sci. USA 101:11571-11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngugi, D. K., M. K. Tsanuo, and H. I. Boga. 2005. Rhodococcus opacus strain RW, a resorcinol-degrading bacterium from the gut of Macrotermes michaelseni. Afr. J. Biotechnol. 4:639-645. [Google Scholar]
- 30.Nordin, K., M. Unell, and J. K. Jansson. 2005. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71:6538-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfennig, N. 1978. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int. J. Syst. Bacteriol. 28:283-288. [Google Scholar]
- 32.Philipp, B., and B. Schink. 1998. Evidence of two oxidative reaction steps initiating anaerobic degradation of resorcinol (1,3-dihydroxybenzene) by the denitrifying bacterium Azoarcus anaerobius. J. Bacteriol. 180:3644-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipp, B., and B. Schink. 2000. Two distinct pathways for anaerobic degradation of aromatic compounds in the denitrifying bacterium Thauera aromatica strain AR-1. Arch. Microbiol. 173:91-96. [DOI] [PubMed] [Google Scholar]
- 34.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178:506-516. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 1989. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Schink, B., B. Philipp, and J. Müller. 2000. Anaerobic degradation of phenolic compounds. Naturwissenschaften 87:12-23. [DOI] [PubMed] [Google Scholar]
- 37.Schmeling, S., A. Narmandakh, O. Schmitt, N. Gad'on, K. Schuhle, and G. Fuchs. 2004. Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186:8044-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnell, S., F. Bak, and N. Pfennig. 1989. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch. Microbiol. 152:556-563. [DOI] [PubMed] [Google Scholar]
- 39.Shingler, V. 2003. Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ. Microbiol. 5:1226-1241. [DOI] [PubMed] [Google Scholar]
- 40.Springer, N., W. Ludwig, B. Philipp, and B. Schink. 1998. Azoarcus anaerobius sp. nov., a resorcinol-degrading, strictly anaerobic, denitrifying bacterium. Int. J. Syst. Bacteriol. 48:953-956. [DOI] [PubMed] [Google Scholar]
- 41.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sütfeld, R., F. Petereit, and A. Nahrstedt. 1996. Resorcinol in exudates of Nuphar lutea. J. Chem. Ecol. 22:2221-2231. [DOI] [PubMed] [Google Scholar]
- 43.Tschech, A., and G. Fuchs. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213-217. [DOI] [PubMed] [Google Scholar]
- 44.Tschech, A., and B. Schink. 1985. Fermentative degradation of resorcinol and resorcylic acids. Arch. Microbiol. 143:52-59. [Google Scholar]
- 45.Vialaton, D., and C. Richard. 2002. Phototransformation of aromatic pollutants in solar light: photolysis versus photosensitized reactions under natural water conditions. Aquat. Sci. 64:207-215. [Google Scholar]
- 46.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. Van Dorsselaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238-1252. [DOI] [PubMed] [Google Scholar]
- 47.Zaborina, O., D. L. Daubaras, A. Zago, L. Xun, K. Saido, T. Klem, D. Nikolic, and A. M. Chakrabarty. 1998. Novel pathway for conversion of chlorohydroxyquinol to maleylacetate in Burkholderia cepacia AC1100. J. Bacteriol. 180:4667-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]