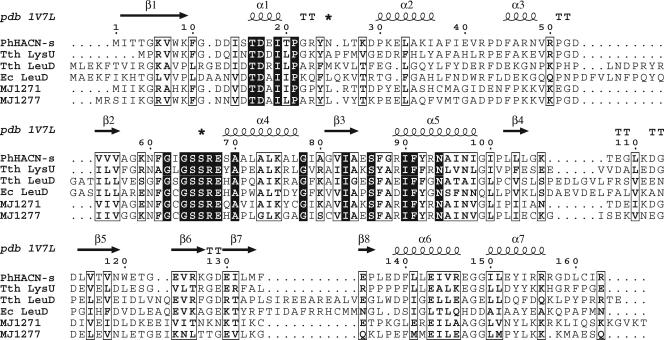
FIG. 2.
Sequence alignment of hydrolyase small subunits shown with the secondary structure of Pyrococcus horikoshii HACN (PhHACN-s; pdb 1V7L) (45). The additional sequences are Thermus thermophilus HACN (Tth LysU; RefSeq accession no. YP_005515.1), T. thermophilus isopropylmalate dehydratase (Tth LeuD; RefSeq accession no. YP_004837.1), Escherichia coli isopropylmalate dehydratase (Ec LeuD; Swiss-Prot accession no. P30126), Methanocaldococcus jannaschii MJ1271 (Swiss-Prot accession no. Q58667), and M. jannaschii MJ1277 (Swiss-Prot accession no. Q58673). Asterisks above the alignment indicate the loop regions that are predicted to bind substrate in the P. horikoshii HACN structure.