Abstract
Cellular iron levels are closely monitored by iron regulatory and sensor proteins of Mycobacterium tuberculosis for survival inside macrophages. One such class of proteins systematically studied in eukaryotes and reported in a few prokaryotes are the iron-responsive proteins (IRPs). These IRPs bind to iron-responsive elements (IREs) present at untranslated regions (UTRs) of mRNAs and are responsible for posttranscriptional regulation of the expression of proteins involved in iron homeostasis. Amino acid sequence analysis of M. tuberculosis aconitase (Acn), a tricarboxylic acid (TCA) cycle enzyme, showed the presence of the conserved residues of the IRP class of proteins. We demonstrate that M. tuberculosis Acn is bifunctional. It is a monomeric protein that is enzymatically active in converting isocitrate to cis-aconitate at a broad pH range of 7 to 10 (optimum, pH 8). As evident from gel retardation assays, M. tuberculosis Acn also behaves like an IRP by binding to known mammalian IRE-like sequences and to predicted IRE-like sequences present at the 3′ UTR of thioredoxin (trxC) and the 5′ UTR of the iron-dependent repressor and activator (ideR) of M. tuberculosis. M. tuberculosis Acn when reactivated with Fe2+ functions as a TCA cycle enzyme, but upon iron depletion by a specific iron chelator, it behaves like an IRP, binding to the selected IREs in vitro. Since iron is required for the Acn activity and inhibits the RNA-binding activity of Acn, the two activities of M. tuberculosis Acn are mutually exclusive. Our results demonstrate the bifunctional nature of M. tuberculosis Acn, pointing to its likely role in iron homeostasis.
Aconitases (Acns), the iron-sulfur proteins, are considered ancient enzymes in the evolution of metabolic pathways (5). The iron-sulfur clusters of these proteins not only participate in electron transport during reversible isomerization of citrate and isocitrate in citric acid cycle (7) but also serve as iron and oxygen sensors of the cell (6, 23). The binary activities are exerted through the assembly and disassembly of iron and sulfur clusters (6, 23). The protein with an intact 4Fe-4S cluster functions as Acn, whereas the protein with 3Fe-4S is an RNA-binding translational regulator (21, 29). The stability and functionality of Acns as translation regulators are affected by not only iron levels but also oxidative stress, which induces these iron-regulatory proteins (IRPs) to bind to iron-responsive elements (IREs) and maintain iron homeostasis (15). IRPs maintain iron homeostasis by posttranscriptionally binding to conserved RNA stem-loop structures or IREs present at either the 5′ or 3′ ends of untranslated regions (UTRs) of mRNA. Depending on whether the IRE is present on the 3′ or 5′ end, binding of IRPs to IREs either protects the mRNA from degradation or inhibits its translation (1, 24). IRE-like sequences are present in the UTRs of at least two enzymes of the citric acid cycle, Acn and succinate dehydrogenase in mammals, implying that IRPs play an important role in mediating iron regulation of mitochondrial energy production (20). IRPs are also activated by both hydrogen peroxide and iron-mediated oxidative stress (27). The reactivity of H2O2 with iron (Fenton reaction) intimately connects oxidative stress and cellular iron metabolism (28). Thus, recruitment of IRPs constitutes a highly effective strategy employed by pathogens for survival.
Acns and IRPs are related with respect to the conserved amino acid residues across the family. This became evident when active-site residues identified in the pig heart mitochondria Acn crystal structure were found to be conserved across mammalian IRPs (16). It is worth mentioning that most of the knowledge on IRP binding to the IRE and the regulatory consequences has been collected from eukaryotic systems where partitioning between cytosolic Acn (IRPs) and mitochondrial Acn exists (references 14, 19, 26, 30, 31 and the references therein). However, only a few Acns have been reported so far from prokaryotes. Based on primary structure similarity, all bacterial Acns, including the α-proteobacterial Acns, are categorized mainly either into the Acn group similar to eukaryotic IRP or cytosolic Acn (AcnA/IRP group) or into the Acn group found only in bacteria (AcnB) (4, 37). Several bacteria, such as Escherichia coli, have two isoforms of Acn, AcnA and AcnB, with different physiological properties and expression profiles (22, 34, 36), while prokaryotes like Bacillus or Xanthomonas (1, 33, 38) have only one Acn. Bacillus Acn has been reported to bind to IRE-like sequences and therefore displays IRP properties (1). The M. tuberculosis energy cycle has separate oxidative and reductive half-cycles (35). M. tuberculosis carries a single copy of the Acn gene (acn) coded by Rv1475c. Sequence comparisons of Mycobacterial Acn with E. coli Acns AcnA and AcnB showed that mycobacterial Acn has a closer identity to AcnA of E. coli (∼60% identity) than to AcnB (∼20% identity). Earlier reports showed that AcnA of E. coli is induced in stationary phase or during oxidative stress (22). The fact that the AcnA/IRP group is less sensitive to oxygen-mediated inactivation (36, 37) makes it logical to argue that aerobic respiration in M. tuberculosis would use the more stable AcnA/IRP-like Acn for energy metabolism, a feature important for survival under oxidative stress. The information that M. tuberculosis Acn expression was downregulated 4.74-fold in a starvation model (8) and upregulated under iron overload conditions (39) suggests that this protein may be associated with both energy and iron metabolism.
In the present study we describe M. tuberculosis Acn as a bifunctional protein, showing enzyme activity when the enzyme is reconstituted by iron and RNA-binding activity when the enzyme is deprived of iron, and we show that these two properties of M. tuberculosis Acn are mutually exclusive. We further describe the functional oligomeric state and basic biochemical properties of M. tuberculosis Acn as a tricarboxylic acid cycle (TCA) cycle enzyme.
MATERIALS AND METHODS
PCR amplification, cloning, and expression of M. tuberculosis Acn. The 2.892-kb open reading frame (ORF; Rv1475c) sequence corresponding to Acn was amplified from H37Rv genomic DNA using proofreading Taq polymerase (AccuTaq LA DNA Polymerase; Sigma) using primers having restriction sites XhoI and HindIII at the 5′ end (forward, ATTCCTCGAGATGTGACTAGCAAATCTGTG; reverse, ATGGAAGCTTTCAGCCTGACTTCAGTATGT). The PCR cycles (35 cycles) were composed of denaturation (94°C for 1 min), annealing (50°C for 1 min), and elongation (68°C for 3.5 min) steps. The insert was cloned in the expression vector pRSET-C at the XhoI/HindIII site, followed by overexpression in E. coli BL21(DE3). The overexpressed His-tagged recombinant protein was purified by Ni2+-nitrilotriacetic acid affinity chromatography. The cells, transformed with the recombinant plasmid, were grown in Terrific broth or as previously described (18), and the expression of the gene was achieved. The cells were lysed by sonication and collected by centrifuging at 16,000 × g for 30 min at 4°C, and the clear lysate was loaded onto the Ni2+-nitrilotriacetic acid column. The column was washed with 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8. The protein was eluted in the same buffer supplemented with 250 mM imidazole. The protein was 90 to 95% pure as checked by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining. The purified protein was dialysed against 20 mM Tris-Cl (pH 8)-100 mM NaCl-3% glycerol and quantified using the Bradford method of protein quantification.
Reconstitution and inactivation of the native protein.
The protein purified under native conditions was reconstituted by incubating with 1 mM ferrous ammonium sulfate and 10 mM dithiothreitol in 50 mM Tris-Cl, pH 8, at 25°C for 20 min. The reaction mixture was centrifuged at 16,000 × g at 4°C for 10 min to pellet any precipitated protein and then was dialysed as above. The purified enzyme was inactivated by the addition of a 0.5 mM concentration of the specific iron chelator dipyridyl or nonspecific metal chelator EDTA whenever required.
Biochemical assays.
Acn activity was measured spectrophotometrically by monitoring the time-dependent conversion of isocitrate to cis-aconitate at 25°C in a Unicam UV/visible light spectrometer at 240 nm. The standard assay solution (400 μl) contained 20 mM triethanolamine chloride buffer, pH 8, 2 mM dl-isocitrate, 100 mM NaCl, and a 30 to 50 nM concentration of the enzyme. Tolerance to pH change by the enzyme was measured by altering the pH of the buffer (range, 5 to 11). The pH dependence of the enzyme was measured using a 20 mM concentration of the following buffers: phosphate buffer (pH 5.7 to 7), Tris-Cl (pH 5 to 9), and CAPS [3-(cyclohexylamino)-1-propanesulfonic acid; pH 9 to 11]. The kinetic parameters were determined by altering the concentration of the substrate (0.1 mM to 4 mM). The values were plotted and counter-checked with a Michaelis-Menten plot of velocity (v) versus substrate concentration ([S]), with a Hanes plot ([S]/v versus [S]), and with a direct linear plot (v versus [S], with median values) for calculating Km and Vmax.
Size exclusion chromatography.
Size exclusion chromatography was performed at room temperature using fast-protein liquid chromatography (FPLC) and a Superdex-200 HR 10/30 column (Amersham Pharmacia Biotechnology). Calibration of the column was performed as described by Chauhan et al.(12), using protein molecular mass standards for gel filtration (Sigma). The column was equilibrated with three bed volumes of the elution buffer prior to each run. Protein elution was monitored at A280. Recombinant proteins at a concentration of 1.2 mg/ml were used for all gel filtration experiments.
Scanning of M. tuberculosis genome for IRE-like sequences.
The M. tuberculosis genome was scanned using the pattern search program of TubercuList (http://genolist.pasteur.fr/TubercuList/) for the sequence CNNNNNCAGUG, with or without a single mismatch in the region CAGUG, located within 200 bp upstream of a start codon or 200 bp downstream of a stop codon (1). The selected sequences were then subjected to secondary structure prediction (http://www.genebee.msu.su/services/rna2_reduced.html) for a possible stem-loop-like configuration. The corresponding deoxynucleotide oligomers for selected IRE-like sequences were commercially synthesized (Microsynth, Switzerland). The selection of putative IRE-like sequences for the binding assays was based on their presence at UTRs of ORFs that were annotated to be involved in iron or oxygen metabolism of the cell. The deoxyribonucleotide sequences corresponding to IRE-like ribonucleotide sequences are given below (IRE-like sequences are underlined and conserved residues are in boldface): 5′ UTR of human ferritin (control), AATTCGGGAGAGGATCCTGCTTCAACAGTGCTTGGACGGATCCA; 3′ UTR of M. tuberculosis trxC (no mismatch; 133 bases downstream to the 3′end), AATTCGGCGATGCGCTGCGCTGTGGCGACCGCAGTGCGGCCGTCACCGAGATCCGGA; 5′ UTR of M. tuberculosis ideR (single mismatch; 62 bases upstream to 5′ end), AATTCGGTAGCAGACGGTATGCCCGCCGCGCCAGCGGCGGGCATACCGCTGCGGTGA.
In vitro transcription.
All RNA work was carried out using diethyl pyrocarbonate-treated water. The complementary oligodeoxyribonucleotides of the selected sequences were cloned in pGEM-3Zf vector at EcoRI and HindIII sites. The vector was linearized using HindIII and used as a template for in vitro transcription using T7 RNA polymerase. For gel shift assays, the 10-μl reaction mixture contained 150 ng of DNA template, 10 mM dithiothreitol, 20 to 25 units of T7 RNA polymerase, 1× RNA polymerase reaction buffer, and a 0.5 mM concentration of ATP, GTP, and UTP with 0.5 mM CTP (whenever unlabeled CTP was used) or 0.05 mM CTP with 20 μCi of [α-32P]CTP (whenever the probe was radiolabeled). The reaction was incubated at 37°C for 1 h followed by RNase-free DNase treatment (1 unit of DNase per reaction) for 15 min at 37°C. After phenol extraction, RNA was precipitated with the addition of 0.5 M ammonium acetate, 15 to 20 μg of Saccharomyces cerevisiae RNA, and 1 volume of isopropanol at −20°C overnight. The RNA was dissolved in nuclease-free water. All of the in vitro transcription experiments were followed by purification of the labeled RNA through a Sephadex G50 column. All in vitro transcribed RNAs were checked on 7 M urea-15% acrylamide gels to confirm that the synthesized RNAs were of correct size. Both labeled and unlabeled RNAs were denatured at 85°C for 5 min, followed by renaturation by slow cooling to allow proper folding before being used for gel shift assays.
Gel retardation assays.
The renatured radiolabeled RNA fragments (∼1 pmol) were allowed to bind to ∼3 μg of purified M. tuberculosis Acn in a binding reaction mixture (20 μl) containing 10 mM Tris-Cl, pH 8, 50 mM KCl, 10% glycerol, and 1 μg of total yeast RNA. The reaction was carried out at room temperature for 30 min. The reaction products were loaded onto a 6% nondenaturing polyacrylamide gel and electrophoresed in TGE buffer (12.5 mM Tris base [pH 8.3], 95 mM glycine, 0.5 mM EDTA) at room temperature. A 0.5 mM concentration of dipyridyl was used whenever M. tuberculosis Acn was inactivated, with an incubation time of 15 min at 25°C. The nonspecific metal chelator EDTA was also used in other experiments as described in the figure legends. For competition experiments, a 75× molar excess of nonradiolabeled specific and nonspecific RNA fragments was used. Titrations were carried out for Acn (from 1 μg to 7 μg), RNA probe (0.5 pmol to 2 pmol), iron (0.5 mM to 2 mM), and dipyridyl (0.1 mM to 1 mM) before a decision was made about the final concentrations to be used in the experiment.
RESULTS
Recombinant M. tuberculosis Acn coded by ORF 1475c is a functionally active TCA cycle enzyme with broad pH tolerance; the functional form in vitro is a monomer.
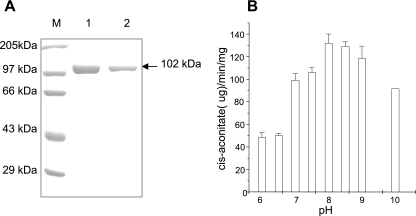
The overexpressed N-terminal His-tagged 102-kDa M. tuberculosis Acn was purified from E. coli BL21(DE3) cells to 90% to 95% homogeneity using a nickel affinity column (Fig. 1A) with a yield of ≥7 mg/750 ml of start culture. Recombinant M. tuberculosis Acn could convert isocitrate to cis-aconitate in a reaction mixture containing 2 mM dl-isocitrate, 100 mM NaCl, and a 30 to 50 nM concentration of the enzyme present in 20 mM triethanolamine chloride buffer, pH 8. The formation of cis-aconitate was monitored spectrophotometrically at 240 nm in a time-dependent fashion at 25°C. The active enzyme was used for further characterization. The activity of M. tuberculosis Acn as a function of pH was studied. Although the optimum pH for the activity of M. tuberculosis Acn, as seen from the profile of pH versus activity, is pH 8, the enzyme remains fairly active within a broad pH range of 7 to 10 (Fig. 1B). The broader activity range of M. tuberculosis Acn is in agreement with its apparent similarity to AcnA of E. coli.
FIG. 1.
(A) Affinity purification of M. tuberculosis Acn. Purified histidine-tagged recombinant protein was checked by 10% SDS-PAGE. Lane M, protein molecular size markers; lanes 1 and 2, purified M. tuberculosis Acn. (B) Profile of pH versus activity of M. tuberculosis Acn. The M. tuberculosis Acn was assayed under different pH conditions as described in the text. The enzyme was active within a broad pH range.
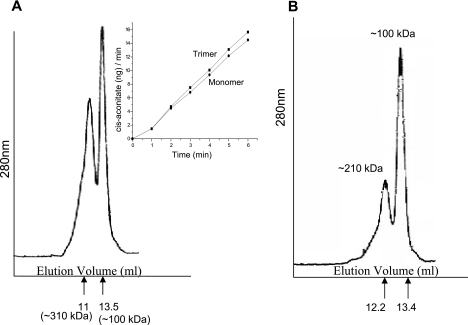
FPLC profiles of M. tuberculosis Acn (Fig. 2A) showed two peaks corresponding to a monomer (∼100 kDa) and trimer (∼310 kDa) in the presence of 20 mM Tris-Cl, pH 8, and 100 mM NaCl. Each fraction was collected separately, quantified, and checked for enzyme activity in a time-dependent fashion (Fig. 2A, inset). The activity curve shown in Fig. 2A (inset) clearly indicates that both the monomer and trimer are enzymatically active. We further calculated the kinetic parameters of the monomer and trimer separately to understand the significance of the trimeric form for the activity of M. tuberculosis Acn. The Km (isocitrate) for the trimer was 0.28 ± 0.08 mM while that for the monomer was 0.56 ± 0.1 mM. Even though an insignificant difference was observed in Km values, the specific activity of both of the forms (150.5 ± 19.4 and 118.1 ± 33.3 μg of cis-aconitate/min/mg of Acn for the trimer and monomer, respectively) remained nearly the same, indicating that the trimer is probably an aggregate of monomers and not a separate oligomeric form.
FIG. 2.
(A) The functional form of M. tuberculosis Acn in vitro is a monomer. The elution profile of native recombinant M. tuberculosis Acn on a Superdex-200 HR 10/30 column showed two peaks corresponding to trimer and monomer. (Inset) Comparative activity of the collected fractions. The specific activities are 150.5 ± 19.4 and 118.1 ± 33.3 μg of cis-aconitate/min/mg of Acn for trimer and monomer, respectively. (B) The trimer of native protein could be disrupted into dimer and monomer upon NaCl treatment. The FPLC profile of 500 mM NaCl-treated M. tuberculosis Acn showed peaks corresponding to dimer and monomer.
To further confirm that the trimer was not a stable functional form of M. tuberculosis Acn and was just an aggregate, we incubated the protein with 300 mM, 500 mM, and 1 mM NaCl for 1 h at room temperature, followed by loading onto a Superdex-200 column for fractionation. It could be clearly seen that the trimer is disrupted into dimer and monomer upon treatment with 500 mM NaCl (Fig. 2B). The protein was subjected to UV cross-linking (data not shown); however, no band corresponding to the size of a trimer could be found upon fractionation by 8% SDS-PAGE. This clearly shows that the trimer is an aggregate formed due to ionic interactions between Acn molecules and does not represent a stable functional form. These biochemical features clearly suggest that M. tuberculosis Acn is possibly a monomeric protein, enzymatically active as a TCA cycle enzyme in converting isocitrate to cis-aconitate at a broad pH range of 7 to 10.
M. tuberculosis Acn binds with high specificity to IRE-like RNA sequences.
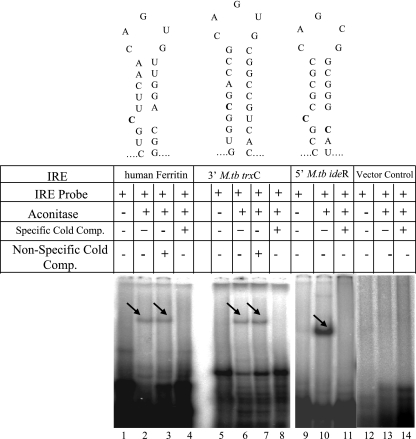
Having shown that M. tuberculosis Acn has enzymatic activity, we carried out electrophoretic mobility shift assays to ascertain if M. tuberculosis Acn displays RNA-binding properties. Binding of purified M. tuberculosis Acn to selected IRE sequences was tested by assaying the interaction between the recombinant protein and in vitro transcribed radiolabeled, 57-nucleotide, RNA carrying IRE-like sequences (Fig. 3). The RNA probes were denatured, which was followed by slow cooling to enable the RNA to attain a proper IRE-like configuration. The presence of an IRE-RNA-Acn protein complex can be clearly seen in electrophoretic mobility shift assays. Compared to the free probe (Fig. 3, lanes 1, 5, 9, and 12), the IRE-RNA-protein complex is evident with the control 5′ human ferritin IRE (Fig. 3, lanes 2 to 4) as well as the selected IRE-like sequences present within the M. tuberculosis genome, i.e., 3′ M. tuberculosis trxC (Fig. 3, lanes 6 to 8) and 5′ M. tuberculosis IdeR (Fig. 3, lanes 10 and 11) under these binding conditions. That these mobility shifts are indeed specific is evident from homologous (specific) and heterologous (nonspecific) cold competition experiments. The labeled IRE-like sequence is effectively competed out in the presence of a 75× molar excess of specific unlabeled RNA corresponding to the ferritin probe (lane 4), 3′ M. tuberculosis trxC (lane 8), and 5′M. tuberculosis IdeR (lane 11), confirming the specificity of the complex. Furthermore, the complex could not be competed out by unlabeled nonspecific RNA (Fig. 3, lanes 3 and 7). Any kind of nonspecific binding with the vector was ruled out by using a plasmid vector RNA control. pGEM-3Zf vector was linearized with HindIII and subjected to in vitro transcription, and the labeled transcript was used as the vector control. M. tuberculosis Acn did not bind to the vector RNA (Fig. 3, lanes 12 to 14). Since all the probes shown are different in Fig. 3 (5′ human ferritin IRE, lanes 1 to 4; 3′ M. tuberculosis trxC, lanes 5 to 8; 5′ M. tuberculosis IdeR, lanes 9 −11; and vector RNA control, lanes 12 to 14), the affinity of M. tuberculosis Acn to bind to these different RNAs varied. However, within a given probe correlation could be seen. For example, in lanes 5, 6, 7, and 8, lane 5 is the free probe, lane 6 and 7 are the complex, and lane 8 is the specific cold competition. A clear correspondence in the intensity of the free probe between lanes 6 and 7 and lane 5 as well as between lanes 6 and 7 and lane 8 can easily be noted. In another control experiment the ability of these RNAs to bind to the nonspecific protein was tested. The selected IRE-like sequences did not bind to any of the nonspecific proteins that were tested, namely, M. tuberculosis isocitrate dehydrogenases ICD-1 and ICD-2 and bovine serum albumin (data not shown). These results demonstrate that M. tuberculosis Acn has RNA-binding activity binding specifically to IRE-like sequences present in UTRs of 3′ M. tuberculosis trxC and 5′ M. tuberculosis IdeR mRNA.
FIG. 3.
M. tuberculosis Acn binds to IRE-like RNA sequences with high specificity. Gel retardation assays were carried out with the control 5′ human ferritin IRE (lanes 1 to 4), 3′ M. tuberculosis trxC (lanes 5 to 8), 5′ M. tuberculosis IdeR (lanes 9 to 11), and vector RNA control (lanes 12 to 14). The renatured radiolabeled RNA fragments were allowed to bind with ∼3 μg of purified M. tuberculosis Acn in a binding reaction (20 μl) containing 10 mM Tris-Cl (pH 8), 50 mM KCl, 10% glycerol, and 1 μg of total yeast RNA. The reaction was carried out at room temperature for 30 min. The specificity of the RNA-protein interaction was checked using 75× molar excess the corresponding specific (lanes 4, 8, 11, and 14) and nonspecific (lanes 3 and 7) competitors. The stem-loop-like secondary structures of the IRE sequences are shown diagrammatically in the figure. M.tb, M. tuberculosis.
RNA-binding activity and Acn activity of M. tuberculosis Acn are mutually exclusive.
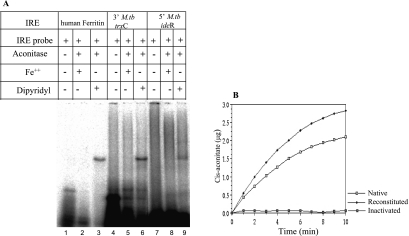
Having shown that M. tuberculosis Acn binds specifically to IRE-RNA, we designed experiments to investigate whether the enzymatic activity is independent of the RNA-binding property. Recombinant purified Acn was reconstituted with iron and inactivated with the specific iron chelator dipyridyl. The reconstituted and the inactivated proteins were checked both for the RNA-binding property (Fig. 4A) and enzyme activity (Fig. 4B). Correlating the activity profiles with the gel shift assays makes it clear that upon treatment with Fe2+, the reconstituted M. tuberculosis Acn becomes more active enzymatically (Fig. 4B) but loses its RNA-binding property (Fig. 4A, lanes 2, 5, and 8). On the other hand, dipyridyl treatment, which specifically chelates iron, inactivates the enzymatic activity of Acn (Fig. 4B) while still preserving its RNA-binding property (Fig. 4A, lanes 3, 6, and 9). These results clearly demonstrate that the ability of M. tuberculosis Acn to act like an RNA-binding protein is independent of its enzymatic property.
FIG. 4.
RNA-binding activity and Acn activity of M. tuberculosis Acn are mutually independent. (A) Gel retardation assays of Fe2+- and dipyridyl-treated M. tuberculosis Acn were carried out to document RNA-protein interactions using 5′ human ferritin IRE (lanes 1 to 3), M. tuberculosis 3′ trxC (lanes 4 to 6), and M. tuberculosis 5′ IdeR (lanes 7 to 9). (B) Comparative enzyme activity curves of native (no iron or dipyridyl treatment), reconstituted (lanes 2, 5, and 8), and dipyridyl-treated (lanes 3, 6, and 9) M. tuberculosis Acn. The reaction conditions shown in the gel retardation assays in panel A were used for enzyme activity determination. M.tb, M. tuberculosis.
DISCUSSION
Postgenomic research efforts have unraveled the functional promiscuity of several M. tuberculosis proteins, in that many have been shown to have additional or new features beyond those originally annotated (3, 10, 11, 13, 32). This work is yet more evidence of such promiscuity shown by M. tuberculosis proteins, since we show for the first time that the M. tuberculosis TCA cycle enzyme Acn not only participates in energy metabolism but also has an RNA-binding property in the context of iron availability. While functioning as a TCA cycle enzyme, M. tuberculosis Acn catalyzes the reversible isomerization of citrate to isocitrate via the intermediate cis-aconitate. On the other hand, as an RNA-binding protein, it binds to selected IRE-like sequences present within the UTRs of 3′ M. tuberculosis trxC and 5′ M. tuberculosis IdeR mRNA. We also demonstrate that these two functions are mutually exclusive. Size exclusion chromatography followed by biochemical characterization of the eluted fractions presented some interesting features of M. tuberculosis Acn. Even though, similar to monomeric proteins of the Acn family, a functional trimeric form was consistently present in gel filtration assays, it is likely that the M. tuberculosis Acn exists as a monomer. This was evident from the dissociation of the trimer into monomer under high-salt concentrations. The observed trimeric form was more likely due to aggregation involving ionic interactions in vitro. While it is difficult to determine enzymatically whether the physiological form of Acn in the cell is a monomer or trimer, what is certain is that the specific activity of the two forms is almost the same (Fig. 2A and B), and similar aggregation in vivo, if any, will not affect their activity. M. tuberculosis Acn could tolerate a broad range of pH values (Fig. 1B), which indicates its robustness, like E. coli AcnA. E. coli AcnA is expressed during stationary phase when the intracellular pH is expected to vary over a wider range than log phase (22). As M. tuberculosis has to survive under very hostile conditions inside human macrophages, it is quite logical that its energy cycle enzymes would have broad tolerance ranges (2). The M. tuberculosis energy cycle has been currently shown to deviate from conventional TCA cycles with a possibility of separate oxidative and reductive half-cycles (35).
The selection of probable IRE-like sequences in the M. tuberculosis genome was difficult because, even though IRPs bind to the consensus IRE sequence with high affinity, earlier studies have shown that IRPs also bind to alternate ligands with relatively different efficiencies. Relative binding efficiency differs in accordance with changes in nucleotides within the stem-loop (9). Phylogenetic relativity, mutational analysis, and comparisons among functional IREs in different transcripts have permitted the identification of the signatures of the IREs that are necessary for high-affinity binding by the IRPs (9). Therefore, two sequences were selected to study the RNA-binding property of M. tuberculosis Acn: the 3′ M. tuberculosis trxC that has no mismatch with the conserved CAGUG sequence and the 5′ M. tuberculosis IdeR that has a single mismatch. Despite a single mismatch in the selected 5′ M. tuberculosis IdeR, both of the selected sequences had a structure resembling the “loop with a neck” that is typical of IREs (diagrammatically shown in Fig. 3). Also, as mentioned earlier, these putative IRE-like sequences are present at the UTRs of ORFs that are annotated to be involved in either iron or oxygen metabolism of the cell.
It is known that the Acn enzyme has three Fe atoms directly bound to the cysteine residues of the enzyme backbone, while the fourth Fe is ligated to the sulfur of the inactive (3Fe-4S) cluster. The Fe is highly prone to dissociation from the cluster (25). The increase in the enzymatic activity of M. tuberculosis Acn (Fig. 4B) is therefore due to the restoration of this labile iron after reconstitution. We also observed that iron and dipyridyl had an antagonistic effect on enzymatic activity and the RNA-binding ability of M. tuberculosis Acn. The reconstituted enzyme did not bind to the IRE-like sequences studied, while inactivation by dipyridyl did not affect the binding affinity of M. tuberculosis Acn (Fig. 4A). Thus, M. tuberculosis Acn could act both as an enzyme and an RNA-binding protein, depending upon the availability of iron to the protein. Binding of M. tuberculosis Acn to such unrelated IREs like that of mammalian ferritin and Bacillus qoxD (data not shown) validates earlier suggestions that such interactions are not species specific, even though the affinity by which interspecies IRE and IRP bind might differ (1, 17, 40). Binding of M. tuberculosis Acn to IRE-like sequences within the M. tuberculosis genome indicates its role in regulating gene expression at a posttranscriptional level. Even though we have established M. tuberculosis Acn as an RNA-binding protein, the mechanism by which it regulates other ORFs remains elusive. It would be important to characterize the Acn regulatory system of M. tuberculosis and its involvement in virulence, identify other Acn-regulated genes and the effects of oxidative stress, and define the molecular basis of the M. tuberculosis Acn and IRE-mRNA interactions.
Acknowledgments
This work was supported by research grants to S.E.H. from the Council of Scientific and Industrial Research and Department of Biotechnology, Government of India. S.B. was supported by a Senior Research Fellowship from CSIR. We thank Shekhar C. Mande (CDFD), for helpful discussions.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Alén, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. USA 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S., A. Nandyala, R. Podili, V. M. Katoch, and S. E. Hasnain. 2005. Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem. 29:6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, S., A. Nandyala, R. Podili, V. M. Katoch, K. J. R. Murthy, and S. E. Hasnain. 2004. Mycobacterium tuberculosis (Mtb) isocitrate dehydrogenases show strong B cell response and distinguish vaccinated controls from TB patients. Proc. Natl. Acad. Sci. USA 101:12652-12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baughn, A. D., and M. H. Malamy. 2002. From the Cover: A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 99:4662-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beinert, H., and M. C. Kennedy. 1993. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 7:1442-1449. [DOI] [PubMed] [Google Scholar]
- 6.Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. [DOI] [PubMed] [Google Scholar]
- 7.Beinert, H., M. C. Kennedy, and C. D. Stout. 1996. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96:2335-2374. [DOI] [PubMed] [Google Scholar]
- 8.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 9.Butt, J., H. Y. Kim, J. P. Basilion, S. Cohen, K. Iwai, C. C. Philpott, S. Altschul, R. D. Klausner, and T. A. Rouault. 1996. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc. Natl. Acad. Sci. USA 93:4345-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakhaiyar, P., and S. E. Hasnain. 2004. Defining the mandate of tuberculosis research in a postgenomic era. Med. Princ. Prac. 13:177-184. [DOI] [PubMed] [Google Scholar]
- 11.Chakhaiyar, P., Y. Nagalakshmi, B. Aruna, K. J. R. Murthy, V. M. Katoch, and S. E. Hasnain. 2004. Regions of high antigenicity within the hypothetical PPE_MPTR Rv2608 ORF show a differential humoral response but similar T cell response in various categories of TB patients. J. Infect. Dis. 190:1237-1244. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan, R., and S. C. Mande. 2001. Characterization of Mycobacterium tuberculosis H37Rv alkyl hydroperoxidase AhpC points to the importance of ionic interactions in oligomerization and activity. Biochem. J. 354:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. R. Murthy, V. M. Katoch, and S. E. Hasnain. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 71:6338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciesla, J. 2006. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim. Pol. 53:11-32. [PubMed] [Google Scholar]
- 15.Fillebeen, C., and K. Pantopoulos. 2002. Redox control of iron regulatory proteins. Redox Rep. 7:15-22. [DOI] [PubMed] [Google Scholar]
- 16.Frishman, D., and M. W. Hentze. 1996. Conservation of aconitase residues revealed by multiple sequence analysis: implications for structure/function relationships. Eur. J. Biochem. 239:197-200. [DOI] [PubMed] [Google Scholar]
- 17.Gegout, V., J. Schlegl, B. Schlager, M. W. Hentze, J. Reinbolt, B. Ehresmann, C. Ehresmann, and P. Romby. 1999. Ligand-induced structural alterations in human iron regulatory protein-1 revealed by protein footprinting. J. Biol. Chem. 274:15052-15058. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, S., S. Rasheedi, S. S. Rahim, S. Banerjee, R. K. Choudhary, P. Chakhaiyar, N. Z. Ehtesham, S. Mukhopadhyay, and S. E. Hasnain. 2004. A novel method for enhancing solubility of the expressed proteins in E. coli. BioTechniques 37:418-424. [DOI] [PubMed] [Google Scholar]
- 19.Gourley, B. L., S. B. Parker, B. J. Jones, K. B. Zumbrennen, and E. A. Leibold. 2003. Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J. Biol. Chem. 278:3227-3234. [DOI] [PubMed] [Google Scholar]
- 20.Gray, N. K., K. Pantopoulos, T. Dandekar, B. A. C. Ackrell, and M. W. Hentze. 1996. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements Proc. Natl. Acad. Sci. USA 93:4925-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haile, D. J., T. A. Rouault, C. K. Tang, J. Chin, J. B. Harford, and R. D. Klausner. 1992. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc. Natl. Acad. Sci. USA 89:7536-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan, P. A., Y. Tang, A. J. Bradbury, A. J. Thomson, and J. R. Guest. 1999. Biochemical and spectroscopic characterization of Escherichia coli aconitases (AcnA and AcnB). Biochem. J. 344:739-746. [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, D. K., J. Jeong, S. K. Drake, N. B. Wehr, T. A. Rouault, and R. L. Levine. 2003. Iron regulatory protein 2 as iron sensor. Iron dependent oxidative modification of cysteine. J. Biol. Chem. 278:14857-14864. [DOI] [PubMed] [Google Scholar]
- 24.Koeller, D. M., J. L. Casey, M. W. Hentze, E. M. Gerhardt, L. N. Chan, R. D. Klausner, and J. B. Harford. 1989. A cytosolic protein binds to structural elements within the iron regulatory region of the transferrin receptor mRNA. Proc. Natl. Acad. Sci. USA 86:3574-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauble, H., M. C. Kennedy, H. Beinert, and C. D. Stout. 1994. Crystal structures of aconitase with trans-aconitate and nitrocitrate bound. J. Mol. Biol. 237:437-451. [DOI] [PubMed] [Google Scholar]
- 26.Muckenthaler, M., N. Gunkel, D. Frishman, A. Cyrklaff, P. Tomancak, and M. W. Hentze. 1998. Iron-regulatory protein-1 (IRP-1) is highly conserved in two invertebrate species: characterization of IRP-1 homologues in Drosophila melanogaster and Caenorhabditis elegans. Eur. J. Biochem. 254:230-237. [DOI] [PubMed] [Google Scholar]
- 27.Nunez, M. T., C. Nunez-Millacura, V. Tapia, P. Munoz, D. Mazariegos, M. Arredondo, P. Munoz, C. Mura, and R. B. Maccioni. 2003. Iron-activated iron uptake: a positive feedback loop mediated by iron regulatory protein 1. Biometals 16:83-90. [DOI] [PubMed] [Google Scholar]
- 28.Pantopoulos, K., and M. W. Hentze. 1995. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 14:2917-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouault, T., and R. Klausner. 1997. Regulation of iron metabolism in eukaryotes. Curr. Top. Cell. Regul. 35:1-19. [DOI] [PubMed] [Google Scholar]
- 30.Rouault, T. A. 2002. Post-transcriptional regulation of human iron metabolism by iron regulatory proteins. Blood Cells Mol. Dis. 29:309-314. [DOI] [PubMed] [Google Scholar]
- 31.Rouault, T. A. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2:406-414. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqi, N., R. Das, N. Pathak, S. Banerjee, N. Ahmed, V. M. Katoch, and S. E. Hasnain. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a TAP-like efflux pump. Infection 32:109-111. [DOI] [PubMed] [Google Scholar]
- 33.Somerville, G., C. A. Mikoryak, and L. Reitzer. 1999. Physiological characterization of Pseudomonas aeruginosa during exotoxin A synthesis: glutamate, iron limitation, and aconitase activity. J. Bacteriol. 181:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, Y., and J. R. Guest. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145:3069-3079. [DOI] [PubMed] [Google Scholar]
- 35.Tian, J., R. Bryk, M. Itoh, M. Suematsu, and C. Nathan. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. USA 102:10670-10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varghese, S., Y. Tang, and J. A. Imlay. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walden, W. E. 2002. From bacteria to mitochondria: aconitase yields surprises. Proc. Natl. Acad. Sci. USA 99:4138-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, T. J., N. Bertrand, J. L. Tang, J. X. Feng, M. Q. Pan, C. E. Barber, J. M. Dow, and M. J. Daniels. 1998. The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol. Microbiol. 28:961-970. [DOI] [PubMed] [Google Scholar]
- 39.Wong, D. K., B. Y. Lee, M. A. Horwitz, and B. W. Gibson. 1999. Identification of fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, D., G. Dimopoulos, A. Wolf, B. Minana, F. C. Kafatos, and J. J. Winzerling. 2002. Cloning and molecular characterization of two mosquito iron regulatory proteins. Insect Biochem. Mol. Biol. 32:579-589. [DOI] [PubMed] [Google Scholar]