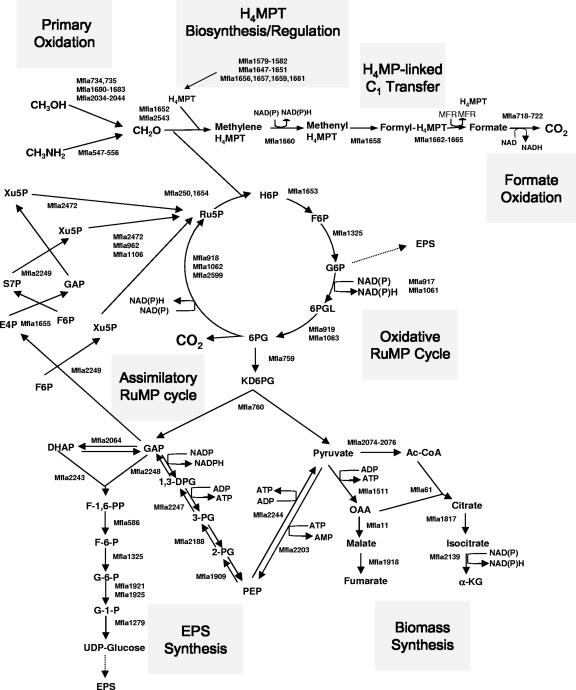
FIG. 1.
Central metabolism of M. flagellatus as deduced from the genome sequence and prior genetic/physiological studies. Gray boxes indicate specific methylotrophy metabolic modules. Enzymes predicted to be responsible for specific reactions are represented by Mf1a numbers of open reading frames as translated from the genome sequence (GenBank accession number NC 007947). CH3OH, methanol; CH3NH2, methylamine; CH2O, formaldehyde; H6P, hexulose 6-phosphate; F6F, fructose 6-phosphate; G6P, glucose 6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose 5-phosphate; Xu5P, xylulose 5-phosphate; E4P, eritrose 4-phosphate; S7P, sedoheptulose 7-phosphate; DHAP, dihydroxyacetone phosphate; F1,6PP, fructose 1,6-bisphosphate; G1P, glucose 1-phosphate; 1,3DPG, 1,3 diphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; α-KG, alpha-ketoglutarate. For more details on methylotrophy genes, see Table S1 in the supplemental material.