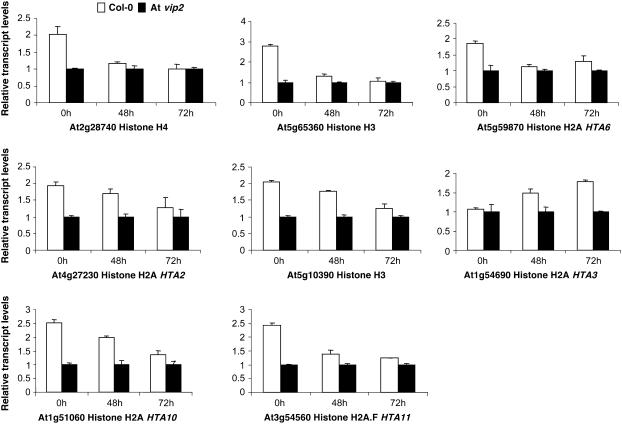
Figure 8.
Validation of the Microarray Data by Real-Time qRT-PCR.
Eight different histone genes that had less transcript abundance in At vip2 compared with Col-0, based on microarray experiments, were selected for validation. Total RNA was extracted from leaves of wild-type Col-0 and At vip2 following agroinfiltration at 0, 48, and 72 h. The first-strand cDNA was synthesized and used for qRT-PCR using gene-specific primers (see Methods). The amount of elongation factor-1-α transcripts was determined and used for normalization. cDNA extracted from Col-0 and At vip2 at 0 HAI was used as calibrator to obtain the relative transcript levels for each gene following agroinfiltration. The data represent the average of three biological replicates, including three technical replicates for each biological replicate with se values shown as error bars.