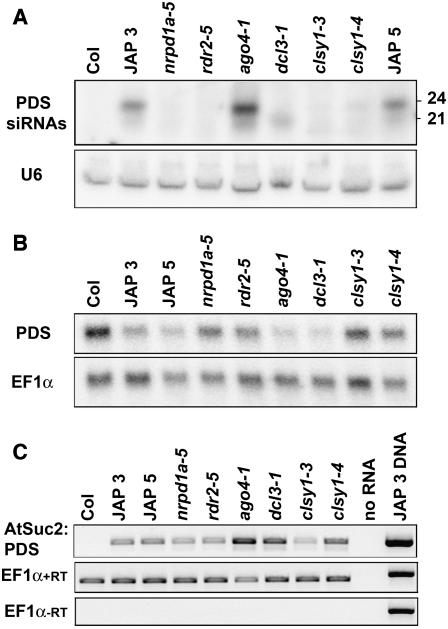
Figure 5.
PDS and PSuc2:PDS RNAs in Wild-Type and Mutant Plants.
(A) Detection of PDS siRNAs in mutant backgrounds. Low molecular weight RNA gel blot analysis was performed on 50 μg of total nucleic acids isolated from the aerial portions of 1-month-old plants. The PDS probe was prepared by T7 transcription from a T7:PDS template amplified from genomic DNA using primers T7 PDS 5′ and PDS 5862r. The U6 signal is included as a loading control.
(B) PDS mRNA levels in 24-nucleotide siRNA pathway mutants. A total of 1.75 μg of poly(A) RNA from 1-month-old plants was analyzed by high molecular weight RNA gel blot. A randomly primed probe from a PDS PCR product (amplified using primers PDS left and PDS right) was used to detect PDS transcript. EF1α was probed as a loading control.
(C) RT-PCRs prepared with the Qiagen one-step RT-PCR kit on total RNA from 1-month-old seedlings. The bottom panel shows a negative control of EF1α RT-PCRs in which the reverse transcription step was omitted from the protocol. Thirty-four cycles of amplification were used for the transgene, and 26 cycles of amplification were used for EF1α.