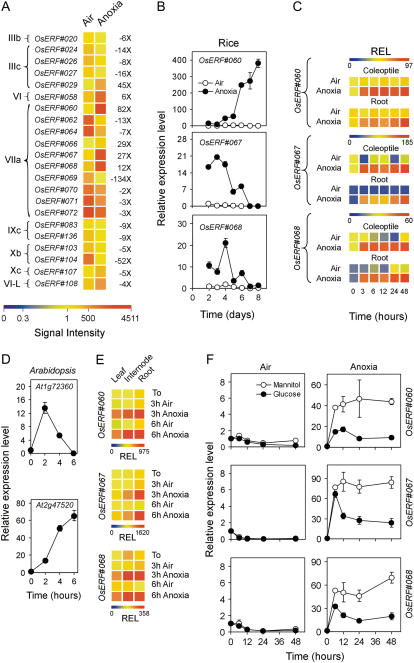
Figure 6.
Effects of anoxia on ERF gene expression. A, Pattern of expression of ERF genes differentially regulated by anoxia; data are expressed as a heat map showing the signal intensity and fold change from the microarray experiment (data are means of two GeneChip experiments) performed on the coleoptiles from rice seeds germinated in air and anoxia for 4 d. ERF categories (left side of the heat map) are as described by Nakano et al. (2006). Fold change is shown on the right side of the heat map (FDR P value < 0.01). See Nakano et al. (2006) or Supplemental Table S2 for corresponding TIGR gene codes. B, Pattern of expression of the ERF genes in coleoptiles from rice seeds germinated in air and anoxia; relative expression level (REL), measured by real-time reverse transcription (RT)-PCR, is shown (REL, 1 = expression data from the aerobic coleoptile at day 2); data are means of three replicates ±sd. C, Pattern of expression of rice ERF genes in the coleoptiles and roots from rice seeds germinated in air for 4 d and then transferred to anoxia for an additional 48-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). D, Pattern of expression of the ERF genes in Arabidopsis seedlings (4-d-old seedlings germinated in air and then transferred to anoxia for up to 6 h). REL, measured by real-time RT-PCR, is shown in the graphs (REL, 1 = expression data from the Arabidopsis aerobic seedlings at t = 0); data are means of three replicates ±sd. E, Pattern of expression of rice ERF genes in the leaves, internodes, and roots from rice plants grown in air for 20 d and then transferred to anoxia for an additional 3- to 6-h period; REL, measured by real-time RT-PCR, is shown as a heat map (REL, 1 = expression data from the aerobic leaf at the beginning of the experiment, t = 0); data are means of three replicates (see Supplemental Table S3 for ±sd values). F, Effects of Glc (100 mm) and mannitol (100 mm, used as an osmotic control) on the expression of rice ERF genes in coleoptiles dissected from 4-d-old aerobically germinated seedlings; REL, measured by real-time RT-PCR, is shown in the graph (REL, 1 = expression data from the aerobic coleoptile at the beginning of the experiment, t = 0); data are means of three replicates ±sd.