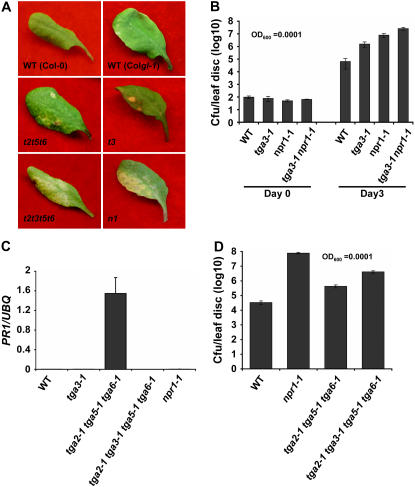
Figure 2.
TGA3 is a positive regulator of PR1 expression and basal resistance. A, Disease symptoms from representative leaves of 4-week-old plants. Infections were carried out using P. syringae pv maculicola ES4326 OD600 = 0.0001. Photographs were taken 3 d postinfection. The experiment was repeated three times with similar results. WT, Wild type; t3, tga3-1; n1, npr1-1; t2t5t6, tga2-1 tga5-1 tga6-1; t2t3t5t6, tga2-1 tga3-1 tga5-1 tga6-1. B, Bacterial titers in the leaves of 4-week-old plants infiltrated with Pseudomonas as described in A. Samples were collected at 0 and 3 d postinfection. Values were averaged from eight samples per genotype and error bars represent se. The experiment was repeated at least three times with similar results. WT, Wild type. C, Basal level expression of PR1 in wild type, tga3-1, tga2-1 tga5-1 tga-6-1, tga2-1 tga3-1 tga5-1 tga6-1, and npr1-1 in the absence of INA. RNA was extracted from 2-week-old seedlings grown on Murashige and Skoog plates. Transcript levels were quantified by real-time RT-PCR and y axis values were normalized to UBQ5 expression. Error bars represent se from three PCR reactions. The experiment was repeated at least three times with similar results. UBQ, Ubiquitin. D, Bacterial titers in the leaves of 4-week-old plants infiltrated with P. syringae pv maculicola ES4326 OD600 = 0.0001. Samples were collected 3 d postinfection. Values were averaged from 16 samples per genotype and the error bars represent se. The experiment was repeated at least three times with similar results.