Abstract
Cell fate determination is often the outcome of specific interactions between adjacent cells. However, cells frequently change positions during development, and thus signaling molecules might be synthesized far from their final site of action. Here, we analyze the regulation of the torso-like gene, which is required to trigger Torso receptor tyrosine kinase activation in the Drosophila embryo. Whereas torso is present in the oocyte, torso-like is expressed in the egg chamber, at the posterior follicle cells and in two separated groups of anterior cells, the border cells and the centripetal cells. We find that JAK/STAT signaling regulates torso-like expression in the posterior follicle cells and border cells but not in the centripetal cells, where torso-like is regulated by a different enhancer. The border and centripetal cells, which are originally apart, converge at the anterior end of the oocyte, and we find that both groups contribute to trigger Torso activation. Our results illustrate how independently acquired expression of a signaling molecule can constitute a mechanism by which distinct groups of cells act together in the activation of a signaling pathway.
Keywords: cell signaling, Drosophila, JAK/STAT
Cell fate in development is often impinged by signaling interactions with neighboring cells. In this regard, a key element is the spatial distribution of signaling molecules in a precise group of cells. This can be achieved either by the regulated expression of the signaling molecule itself or by the restricted activity of a modifying enzyme transforming an inactive precursor into a fully active signaling molecule. In addition, cells can change their neighbors, and thus those molecules might be synthesized far from their final site of action. This is the case for the specification of the most anterior and posterior regions of the Drosophila embryo by the Torso (Tor) receptor tyrosine kinase signaling pathway. The Tor receptor, which is present over the entire blastoderm membrane, is only activated at the poles by a still unknown mechanism that is thought to involve the cleavage of Trunk (Trk), the putative Tor ligand. This mechanism of Tor activation is triggered by the expression of torso-like (tsl), which encodes a protein of unknown function thought to be required for the processing of Trk. Indeed, it is the restricted expression of tsl that determines the localized domain of Tor activation (reviewed in ref. 1).
tsl is expressed in three cell populations in the egg chamber (2, 3) (see Fig. 1). One of them is a group of follicle cells at the posterior end of the egg chamber [named thereafter the posterior follicle cells (PFCs)] that are in close contact with the posterior end of the oocyte, one of the regions where the Tor pathway will be activated upon fertilization. At the anterior end, tsl is expressed in two cell populations, the border cells (BCs) and the centripetal cells (CCs). The BCs arise at the anterior of the egg chamber, far from the oocyte. The CCs are specified later, at an anterior–lateral position close to the oocyte. In contrast with the situation at the posterior end, both the BCs and the CCs are originally distant from the anterior region of the oocyte where the Tor pathway will be activated after fertilization, and it is only after their migration that they become juxtaposed to this region of the oocyte (for a review on the egg chamber and its cell types, see refs. 4 and 5).
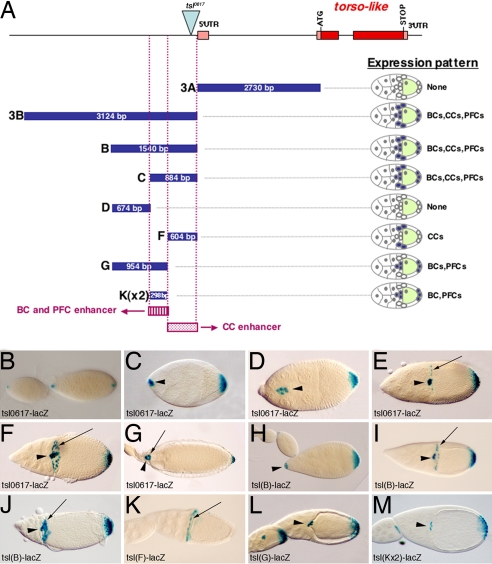
Fig. 1.
Two independent enhancers regulate tsl expression in different cell populations during oogenesis. (A) The tsl genomic fragments tested for enhancer activity by fusion to lacZ reporters. Pink rectangles indicate untranslated exons. Red rectangles indicate translated exons. The blue triangle indicates the P-element in tsl0617. Blue rectangles indicate genomic fragments used for reporter constructs; their size is indicated. Purple rectangles show the identified enhancers. None, No expression. (B–M) X-Gal staining of lacZ reporters. (B–G) tsl0617-lacZ. (B) Expression at stages 7 and 8 of oogenesis in the polar cells at both ends of the egg chamber. (C) At stage 9, expression is first detected in additional PFCs and then in the BCs. (D) At stage 10, expression is detected in the migrating BCs and PFCs. (E) By stage 10B, BCs have reached the oocyte and expression starts to be detected at the CCs. (F) Expression at stage 11. (G) Expression at stage 14. (H–J) tsl(B)-driven expression reproduces all of the tsl pattern [stage 9 (H); stage 10B (I); stage 11 (J)]. (K) tsl(F) drives expression in the CCs. (L) tsl(G) drives expression in the BCs and PFCs. (M) Two copies of the tsl(K) fragment are enough to drive expression in the BCs and PFCs. (K–M) are egg chambers at stages 10B/11. (H–M) These images show ovaries from flies carrying two copies of the construct. Arrows point to CCs, and arrowheads point to BCs. Here and in all images, anterior is to the left.
In this study, we analyze the regulation of tsl expression to find how these distinct groups of follicle cells acquire the ability to express a common signaling factor. We find that JAK/STAT signaling, which is responsible for initially patterning the egg chamber epithelium (6, 7), also regulates tsl expression in the PFCs and BCs. However, we find that this pathway does not regulate tsl expression in the CCs, which is regulated by a different enhancer. We also analyze the functional significance of tsl expression in the BCs and CCs, and find that both groups contribute to trigger Tor activation at the anterior embryonic region. Our results illustrate how independently acquired expression of a signaling molecule in distinct group of cells can constitute a mechanism by which they act together in the activation of a signaling pathway.
Results
Distinct Enhancers Regulate tsl Expression in Specific Groups of Follicle Cells.
Although tsl is expressed in three different groups of follicle cells, these cells are not completely unrelated. Thus, for example, both the BCs and the CCs are derived from a common pool of anterior follicle cells and express and require some of the same genes for their development (8–10). Likewise, many similarities have also been recognized between the BCs and the PFCs (11). This raises the possibility that a common mechanism could single out these cells for tsl expression. Alternatively, each of these groups of follicle cells could be independently targeted to express tsl. As a first attempt to address how these distinct groups of follicle cells acquire the ability to express a common signaling factor, we undertook an analysis of the tsl promoter.
As a first indication of what constitutes the tsl regulatory region, we knew that the P-element insertion carrying the lacZ gene upstream of the 5′-UTR exon in the tsl0617 mutant, thereafter tsl0617-lacZ (2), reproduced all of the features of tsl expression in the follicle cells, as judged by comparison with the tsl in situ hybridization pattern (2, 3) (Fig. 1 B–G). By transformation of lacZ reporter constructs using different regions upstream of the coding sequences of tsl, we found that a single fragment of ≈1,500 bp upstream of the 5′-UTR exon (fragment B in Fig. 1) reproduces the tsl wild-type pattern (Fig. 1 H–J). Further dissection allowed us to split the tsl promoter into two nonoverlapping regions responsible for a different subset of the tsl expression pattern. In particular, we found that a 604-bp sequence (fragment F in Fig. 1) drives expression only in the CCs (Fig. 1K), hereafter referred to as the CC enhancer, whereas an adjacent 954-bp sequence (fragment G in Fig. 1) drives expression in both the BCs and the PFCs (Fig. 1L). Comparison between the different constructs suggested that the enhancer for BCs and PFCs could be further refined to a region of 298 bp (fragment K in Fig. 1). We confirmed this assumption by establishing that two copies of fragment K are sufficient to drive expression in BCs and PFCs (Fig. 1M), hereafter referred to as the BC/PFC enhancer. Thus, in summary, two different regions of the tsl promoter are responsible for distinct subsets of tsl expression. It is remarkable that a single promoter fragment (fragment K) drives tsl expression in two independent group of follicle cells (the BCs and PFCs), whereas separate enhancers (fragments K and F) are responsible for tsl expression in the BCs and CCs, which are derived from a common pool of anterior follicle cells.
The JAK/STAT Pathway Regulates tsl Expression in the BCs and PFCs but Not in the CCs.
In a next step, we addressed what are the mechanisms responsible for tsl expression. The promoter analysis presented above suggested that tsl would probably be regulated by a common mechanism in BCs and PFCs. A likely candidate for such a regulator is the JAK/STAT pathway, which is thought to establish a symmetrical prepattern onto the follicular epithelium by its specific activation at two groups of follicle cells, one at each end of the egg chamber (6, 7). The localized activation of the JAK/STAT pathway could therefore account for the activation of tsl expression in the BCs and PFCs.
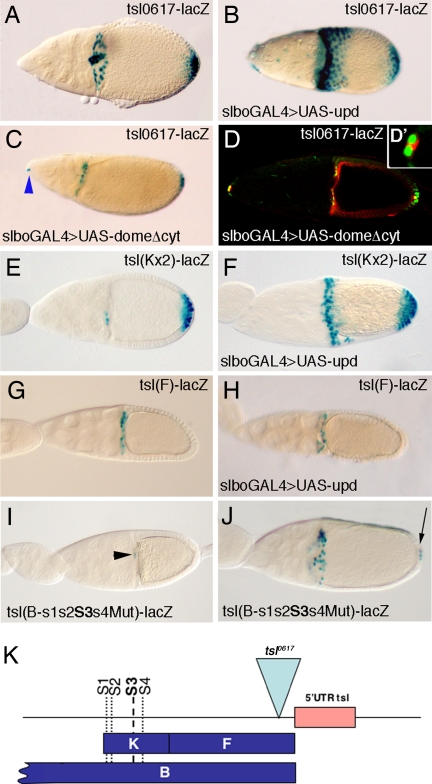
A series of experiments confirmed that the JAK/STAT pathway is responsible for the positive regulation of the BC/PFC enhancer. First, overactivation of the JAK/STAT pathway, by overexpression of its ligand unpaired (upd) with a slboGAL4 driver line, results in an enhancement of tsl0617-lacZ expression (Fig. 2 A and B). slboGAL4 is specifically expressed in the BCs and CCs, and at lower levels in some PFCs (12) (see Figs. 3C and 4A). Accordingly, the overexpression of tsl0617-lacZ is more conspicuous in the regions of higher slboGAL4 expression. Second, impairment of the JAK/STAT pathway, by the widely used mechanism of overexpressing a dominant-negative form of its receptor domeless (dome) (13), with the same slboGAL4 driver, hinders tsl0617-lacZ expression in the BCs (Fig. 2 C and D). Among the BC cluster, we only detect tsl0617-lacZ expression in the inner pair of polar cells (Fig. 2 D and D′), which are a specialized couple of cells that recruit their neighbors to form the BC cluster (14). Expression of tsl0617-lacZ in the polar cells when UASdomeDN is driven by slboGAL4 is consistent with the fact that this driver is expressed in all of the BCs except the polar cells (15) and can be used as an internal control for the experiment. In PFCs, there is a clear reduction of tsl-lacZ expression on domeDN expression, although it is not completely eliminated, most probably because of the lower levels of slboGAL4 expression in the PFCs (12). Importantly, whereas slboGAL4 is very highly expressed in the CCs, overexpression of domeDN does not affect expression of tsl0617-lacZ in these cells, suggesting that an alternative regulatory mechanism of tsl expression is at play in the CCs. Finally, and consistent with the previous observations, we found that overactivation of the JAK/STAT pathway, by expression of upd with slboGAL4, drives ectopic expression of lacZ by the BC/PFC enhancer (Fig. 2 E and F) and has no effect on the CC enhancer (Fig. 2 G and H). These results confirm that the JAK/STAT pathway acts as a primary regulator of tsl expression in the BCs and PFCs.
Fig. 2.
The JAK/STAT signaling pathway regulates tsl expression in the BCs and PFCs. (A–C) Expression of tsl0617-lacZ (A) upon overexpression of upd with the slboGal4 driver (B) and upon overexpression of domeDN with slboGal4 (C), which represses tsl0617-lacZ expression in the BCs and PFCs but not in the CCs. Blue arrowhead in C points the anterior polar cells, which fail to migrate. (D and D′) Immunostaining showing that the most anterior positive cells correspond to the anterior polar cells, where the slboGAL4 driver is not expressed. Green, β-gal; red, Fasciclin III, which accumulates at the contact surface between the two polar cells. (E and F) Overexpression of upd with the slboGAL4 driver activates the BC/PFC enhancer. (G and H) However, overexpression of upd does not activate the CC enhancer. (I and J) Expression driven by a tsl(B) construct with mutations in the four putative STAT binding sites found at the BC/PFC enhancer. At stage 9, no expression can be detected, and at stage 10, only weak expression is seen in the BCs (black arrowhead) but not at the PFCs (I). On the contrary, expression in the CCs is not affected, as shown at stage 11, when occasionally some PFCs express low levels of lacZ (arrow) (J). Expression at the CCs acts as an internal control for expression level of BCs and PFCs. (K) Schematic representation of the B fragment showing the four putative STAT binding sites of the BC/PFC enhancer; note that the CC enhancer remains unaffected. A–J correspond to stages 10B/11.
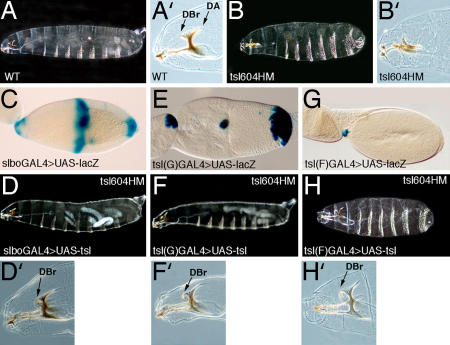
Fig. 3.
tsl overexpression in either the BCs or the CCs is sufficient to trigger Tor receptor activation at the anterior end of the embryo. (A) Cuticle preparation of a WT embryo. (A′) Detail of the WT head skeleton; the dorsal bridge (DBr) and the dorsal arm (DA) are anterior terminal structures. (B) Anterior and posterior terminal structures do not develop in tsl604 homozygous mutant embryos. (B′) The dorsal bridge does not form and the dorsal arm is reduced. (D, F, and H) Overexpression of tsl with the slboGAL4, tsl(G)GAL4 and tsl(F)GAL4 drivers (see pattern of expression in C, E, and G, respectively) rescue the anterior terminal structures of tsl604 mutant embryos; there is also rescue of the posterior terminal structures with the slboGAL4 and tsl(G)GAL4 drivers, which is variable with the slboGAL4 driver. (D′, F′, and H′) Details to show that dorsal bridges are formed at the anterior end using the three drivers.
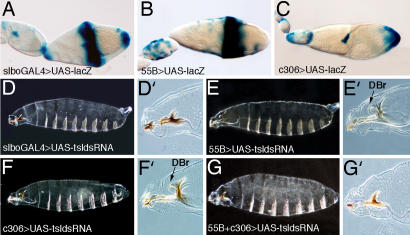
Fig. 4.
RNAi-mediated inactivation of tsl indicates that BCs and CCs contribute to Tor receptor activation at the anterior end of the embryo. (A–C) Pattern of expression of the drivers used to induce the expression of UAStsldsRNA. (D) Expression of UAStsldsRNA with slboGAL4 in both BCs and CCs cause an anterior tsl phenotype. (D′) Detail showing absence of the dorsal bridge. (E) Expression of UAStsldsRNA with the 55B driver does not produce an anterior tsl phenotype. (E′) Detail showing formation of the dorsal bridge (arrow). (F) Similarly, expression of UAStsldsRNA with the C306 driver does not produce an anterior tsl phenotype. Expression of UAStsldsRNA with this driver in the PFCs also gives rise to some embryos with a posterior tsl phenotype. (F′) Detail showing formation of the dorsal bridge (arrow). (G) However, expression of UAStsldsRNA with both the 55B and C306 drivers causes an anterior tsl phenotype. (G′) Detail showing absence of the dorsal bridge. In all images, anterior is to the left and posterior is to the right.
We next investigated whether the effect of the JAK/STAT signaling on tsl expression could be direct. It is known that the JAK/STAT pathway is required for establishment of the BC fate (14, 16, 17). Indeed, BC expression of slbo, a gene coding for a transcription factor, is induced by the JAK/STAT pathway (14, 16). Therefore, the effect of this pathway on tsl expression could be indirect. However, pointing to a direct effect, we detected four putative STAT binding sites (see Methods) in the BC/PFC enhancer (Fig. 2K). Thus, we sought to find whether the JAK/STAT pathway was directly responsible for tsl expression in BCs and PFCs by mutating these putative STAT binding sites in the context of the whole tsl minimal promoter linked to the lacZ reporter. We have found that expression of such a reporter is very much reduced in the BCs and PFCs but remains normal in the CCs (Fig. 2 I and J). The reduction in expression in BCs and PFCs is very strong: at stage 9, no expression is detected, by stage 10 only weak expression can be detected in the migrating BCs when they are close to the oocyte (Fig. 2I), and by stage 11, a few PFCs show very low expression (Fig. 2J). These results demonstrate a key role of these putative STAT binding sites in the regulation of tsl expression in the BCs and PFCs.
The experiments described above show that tsl expression in the CCs is independent of the JAK/STAT pathway activity. As mentioned above, expression of domeDN appears not to affect tsl0617-lacZ expression in the CCs (Fig. 2 C and D). In addition, the tsl CC enhancer does not respond to the overactivation of the JAK/STAT pathway (Fig. 2H). We examined whether other pathways that have been shown to be active in the CCs, such as dpp or Notch (18–20), play a role in CC tsl expression, but did not find a specific effect with these (data not shown). Additionally, we tested factors for which we found putative binding sites in the tsl CC enhancer, such as MyoD or Su(H), but again we failed to see a specific effect (data not shown). In summary, we show that JAK/STAT signaling directly activates BC and PFC expression of tsl. This pathway does not promote tsl expression in the CCs, and the nature of how this expression is achieved remains unknown.
Assessment of tsl Contribution from BCs and CCs to Tor Signaling.
To evaluate the specific contributions of tsl expression in BCs and CCs to Tor signaling at the anterior pole of the embryo, we attempted to alter tsl expression distinctly in these two cell populations. In a first approach, we examined whether tsl expression in either the BCs or the CCs was sufficient to trigger Tor activation in the embryo. To this end, we used the UAS/GAL4 system to drive tsl expression, separately or concurrently, in these groups of cells in otherwise tsl mutant flies. As a positive control, we first confirmed that expression of a UAStsl line in both the BCs and CCs, with a slboGAL4 driver, rescues the tsl phenotype at the anterior end of embryos (Table 1 and Fig. 3D). This same assay rescues the tsl posterior phenotype (Fig. 3D), although not in all of the mutant embryos, probably because of the lower levels of expression of slboGAL4 at the PFCs (12) (Fig. 3C).
Table 1.
Percentage of tsl mutant embryos with anterior terminal structures upon expression of tsl in the BCs and/or the CCs
| Genotype | Expression pattern of the Gal4 lines | Embryos with anterior terminal structures, % | n |
|---|---|---|---|
| UAS-tsl/+; CyO/+; tsl604HM | 0 | 69 | |
| UAS-tsl/+; slboGal4/+; tsl604HM* | BCs, CCs, PFCs | 100 | 121 |
| UAS-tsl/+; tsl(G)Gal4/+; tsl604HM | BCs, PFCs | 100 | 56 |
| UAS-tsl/+; tsl(F)Gal4/+; tsl604HM | CCs (few and late) | 100 | 39 |
Rescue of anterior terminal structures was scored by the complete formation of the dorsal bridge. n, Number of embryos.
*tsl overexpression with the slboGAL4 line generates many embryos with defects in head involution that could not be scored for the formation of the dorsal bridge. Embryos examined for anterior rescue (n = 121 of 218) are those with normal head involution. All experiments were done at 25°C.
To drive tsl expression in the BCs, but not in the CCs, we generated a GAL4 line by using the G fragment, which contains the BC/PFC enhancer from the tsl promoter (Fig. 1). This line [tsl(G)GAL4] combined with a UASlacZ reporter line recapitulates the expression in those cell populations (Fig. 3E). We found that expression of UAStsl with this line also rescues the tsl mutant phenotype both at the anterior end (Table 1 and Fig. 3F) as well as at the posterior end (Fig. 3F). We attempted the same strategy to generate a GAL4 line with the CC enhancer of tsl. The newly generated line [tsl(F)GAL4] was only able to drive expression of a UASlacZ reporter late in oogenesis, and only in a few CCs (Fig. 3G). Nevertheless, expression of UAStsl driven by this line also rescues the mutant phenotype of tsl embryos at their anterior termini (Table 1 and Fig. 3H). Together, these results suggest that tsl expression in either the BCs or the CCs can trigger Tor receptor activation.
In a complementary approach, we tried to selectively inactivate tsl in either the BCs or the CCs. To this end, we generated transgenic flies carrying a UAStsldsRNA construct. We then drove expression of tsldsRNA with follicle-specific GAL4 drivers. As for the rescue experiments, we used the slboGAL4 line to drive expression of the construct in the BCs, CCs, and PFCs. We found that this combination generates a tsl phenotype in the anterior region in 100% of embryos examined, indicating that under this condition RNAi can successfully impair tsl function (Table 2 and Fig. 4D). However, in contrast with the clear effect at the anterior pole, this combination hardly produced any effect at the posterior embryonic region (Fig. 4D), where, as mentioned, the slboGAL4 line is expressed at a lower level (12).
Table 2.
Percentage of embryos with an anterior tsl mutant phenotype upon expression of UAStsldsRNA in the BCs and/or the CCs
| Genotype | Expression pattern of the Gal4 lines | Embryos with anterior tsl mutant phenotype, % | n |
|---|---|---|---|
| slboGal4/+; UAStsldsRNA/+ | BCs, CCs, PFCs | 100 | 85 |
| 55B/UAStsldsRNA | CCs* | 0 | 101 |
| c306/+; UAStsldsRNA/+ | BCs, PFCs† | 0 | 48 |
| c306/+; 55B/UAStsldsRNA | BCs, CCs,* PFCs† | 61 | 87 |
The anterior tslphenotype was scored by absence of the dorsal bridge; in some cases, expression of UAStsldsRNA with slboGAL4 generated embryos with defects in head involution that could not be scored for the formation of the dorsal bridge. Embryos scored are those with normal head involution. n, Number of embryos.
*Broad region around CC from stage 10B, including some stretched cells (see Methods and ref. 31).
Although expression of the UAStsldsRNA construct in both the BCs and the CCs with slboGAL4 produced an anterior tsl phenotype, we were not able to reproduce an anterior tsl phenotype by driving expression of the same construct with several GAL4 lines tested that were specifically expressed either in the BCs or in the CCs (see Methods). One possible reason for this failure to obtain a phenotype could be simply a failure to completely inactivate tsl function in one or the other cell population. An alternative, but not exclusive, possibility may be that tsl expression from one of these groups of cells might compensate for reduction of tsl expression from the other group. Thus, we combined GAL4 lines expressed either in the BCs or the CCs and found one pair (lines C306 and 55B) (Fig. 4 B and C) that, together with the UAStsldsRNA line, produced an anterior tsl phenotype (Table 2 and Fig. 4G). It has to be emphasized that neither line on its own is able to generate an anterior tsl phenotype (Table 2 and Fig. 4 E and F). In particular, we have confirmed that the 55B line is not expressed in the BCs (see Methods), and thus its combined effect with C306 in the RNAi experiment suggests that the anterior tsl phenotype is produced by inactivation of tsl in both the BCs and CCs. It is also important to note that expression of UAStsldsRNA with both C306 and 55B lines produces an anterior tsl phenotype in 60% of the embryos (Table 2), indicating that this combination does not completely inactivate tsl function. However, irrespective of the actual extent of RNAi-mediated inactivation, these results indicate that both the BCs and CCs can contribute to provide sufficient levels of tsl activity to efficiently trigger anterior Tor signaling.
Discussion
One specific feature of Tor pathway signaling is the fact that the cell–cell contact at the origin of the triggering mechanism is established much earlier than the signaling mechanism itself: specific follicle cells expressing tsl contact the oocyte during oogenesis and as a result it is thought that the eggshell is modified at both ends in such a way that much later, with fertilization, the Tor pathway will be activated specifically in those embryonic regions. In this regard, it has been shown that Tsl protein accumulates in the polar regions of the vitelline membrane, suggesting a mechanism for the transfer of the positional information from the follicle cells to the developing embryo (21). Here, we analyzed a second feature of the activation of the Tor pathway: the spatial expression of tsl at the signaling cells.
Patterning by JAK/STAT Signaling: From the Egg Chamber to the Embryo.
We found that tsl expression is controlled by different cis-regulatory regions and different transactivating factors independently in different cell populations: a single promoter fragment responds to JAK/STAT signaling and activates tsl expression in both the BCs and PFCs, whereas another enhancer drives tsl expression in the CCs. Moreover, we have found putative STAT binding sites in the identified BC/PFC enhancer. Mutations in those sites greatly reduce tsl-lacZ expression in the BCs and PFCs, pointing to a direct regulation by the JAK/STAT pathway. The fact that some reporter expression can occasionally be detected in those cells even when these sites are mutated could be attributed to regulation by other factors, which could also contribute to tsl expression in BCs and PFCs. In this regard, microarray analysis has shown that activity of the slbo transcription factor, which has been shown to function as a simple transcriptional activator (22) and whose expression is also dependent on the JAK/STAT pathway (14, 16), induces a 2-fold increase of tsl expression (23, 24). In summary, our results show that the JAK/STAT pathway acts as a primary regulator of tsl expression in the BCs and PFCs.
The JAK/STAT pathway is triggered in the Drosophila egg chamber by localized expression of its ligand, upd, in two polar cells at each end of the chamber (6, 14, 16). Signaling from this pathway is responsible for the patterning of the follicle cells at both ends of the egg chamber (6, 7), and our results show now that it is also responsible for tsl expression in the BCs and the PFCs. Thus, these results indicate that a common mechanism is responsible for initially patterning the egg chamber terminal epithelium and later triggering the mechanism that specifies the embryonic terminal regions.
At the anterior end of the egg chamber, three populations of follicle cells can be distinguished: the BCs, the CCs, and the stretched cells in between. Among those, BCs and CCs, but not stretched cells, express tsl. Although the role of the JAK/STAT pathway in patterning the follicle cells at both ends of the egg chamber is well established, there are conflicting data about whether a gradient of its ligand, upd, could indeed be responsible for patterning all of the anterior follicle cells (7, 16, 25). If that was the case, it might be expected that the JAK/STAT pathway could play a role in tsl expression in both the BCs and the CCs. In this scenario, absence of tsl expression in the stretched cells could be due to specific mechanisms of tsl gene repression in those cells. Conversely, our results show that the JAK/STAT pathway does not have a specific role in the activation of tsl in the CCs. These results do not necessarily argue against a gradient of upd. It could be argued, for example, that lower levels of JAK/STAT signaling in the CCs might not be sufficient to trigger activation of the BC/PFC enhancer. Alternatively, it could also be the case that a specific repressor element in this enhancer might inhibit its expression in the CCs. However, irrespective of a role of the upd gradient in patterning the follicle cells, our results show that tsl expression in the CCs is independent of JAK/STAT. This result indicates that there are JAK/STAT-independent differences within the anterior epithelial cells of the egg chamber, as was previously hypothesized (25).
tsl Expression in Anterior Follicle Cells: Independent Regulation of a Determinant Molecule in Two Cell Populations as a Means to Trigger Cell Signaling.
Our results show that the two groups of anterior cells, the BCs and the CCs, contribute to trigger anterior Tor activation. Moreover, they indicate that this is accomplished by independent regulation of tsl in each of these cell populations. At first glance, either the BCs or the CCs appear to be sufficient to trigger Tor activation. Thus, GAL4-driven expression of tsl in either the BCs or the CCs is able to promote normal development of the terminal anterior structures in embryos derived from otherwise tsl mutant females. Additionally, RNAi-mediated inactivation of tsl in either the BCs or the CCs is not able to generate an anterior tsl phenotype, whereas inactivation in both the BCs and CCs produces embryos with anterior tsl mutant phenotypes. Thus, tsl expression in the BCs and CCs might be redundant. However, there are some caveats to those experiments that should be considered. First, GAL4-driven expression might generate higher tsl levels than the normal in the BCs or CCs. Second, in our experiments, RNAi-mediated inactivation does not completely impair tsl function; this is clearly observed because ≈40% of the embryos develop anterior terminal structures even when UAStsldsRNA is expressed in both the BCs and the CCs using the C306 and 55B drivers (Table 2).
Given these results, we propose that an absolute level of tsl expression may be crucial to trigger Tor signaling. Therefore, it might not be so important whether tsl is supplied by the BCs or the CCs, provided it reaches an absolute amount. This would explain why overexpression of tsl in either the BCs or the CCs can rescue the anterior tsl mutant phenotype. It would also explain the additive effects of lowering tsl activity from the BCs and the CCs to generate an anterior tsl phenotype. Besides, it has to be considered that too much Tsl could also be damaging. In this regard, it has to be noted that tsl overexpression driven by the slboGAL4 driver produces head involution defects in many embryos (Table 1). Taking this into account, expression of tsl from both the BCs and CCs could be a means to reach a minimum amount of Tsl product, but also not to exceed a certain limit.
To understand how such a mechanism could have been established, we should consider the differences in ovary organization among insects. Although all insect ovaries consist of morphologically and physiologically discrete entities (the ovarioles), there are differences on how the oocyte is positioned in reference to the follicle cells. In more ancient insects, the oocyte is surrounded by a monolayer of somatic follicle cells. Conversely, in more evolved insects, a group of nurse cells are clustered at the anterior end of the oocyte and it is only later that the anterior side of the oocyte is separated from the nurse cells and contacts the follicle cells (for a description, see ref. 26). In Tribolium, an insect with a more primitive ovary in which tsl expression has been examined, tsl is precisely expressed in the follicle cells overlying both edges of the oocyte (27). Therefore, Drosophila tsl expression in the BCs and PFCs may represent an adaptation, or the remnant, of a more ancient pattern of tsl expression. The difference in Drosophila is that the anterior tsl-expressing follicle cells, initially separated from the oocyte, have acquired the capacity to migrate through the nurse cells to reach the anterior end of the oocyte. Thus, two insects with different type of ovaries share a common pattern of tsl expression in two groups of follicle cells at both ends of the oocyte, although the mechanism to position these cells next to the oocyte differ in both insects. Conversely, tsl expression in the CCs of Drosophila appears to be a more recent acquisition. The CCs are a new particularly evolved set of follicular cells that migrate to separate the oocyte from the adjacent nurse cells. In this context, concomitant tsl expression in the CCs in Drosophila may have been independently attained by the acquisition of a new distinct enhancer in the tsl promoter.
Therefore, the complex pattern of tsl expression could provide a means to ensure the full triggering and robustness of Tor receptor tyrosine kinase activation and illustrates a mechanism by which the full response of a receptor cell can be accomplished by the independent acquisition of signaling capacity in distinct cell populations and their combined action.
Methods
Drosophila Stocks and Genetics.
We used the following Drosophila stocks described in Flybase (http://flybase.bio.indiana.edu): tsl0617, tsl604, slboGAL4, dppGAL4, UASupd, UASdomeΔcyt, UAStsl, and UASlacZ. Ectopic expression was obtained by using the UAS/GAL4 system (28). For the RNAi-mediated interference, we used the tsl(G)GAL4, 198Y, c306, c522, and cb07 lines expressed in the BCs and the tsl(F)GAL4, cb35, 55B, cb41, and dppGAL4 expressed in the CCs. For a description of these lines, see the following sources: tsl(G)GAL4 and tsl(F)GAL4 (see below), 198Y, c306 and c522 (29), cb07, cb35 and cb41 (30), and 55B (28, 31). We have examined egg chambers from flies carrying the 55B line, a UASGFP construct and the tsl(G)-Bgal construct, which is specific for the BCs and PFCs at the confocal and in no case is there an overlap between the two domains (n = 14) (data not shown). Transgenic lines were generated by P-element transformation (32). Different lines from each construct were analyzed.
Molecular Biology.
tsl-lacZ constructs.
Different fragments from the tsl locus were amplified by PCR using primers with 5′ overhang specific restriction enzyme sites and cloned into the C4PLZ plasmid (33). Details of cloning can be provided on request.
The putative STAT binding sites in the BC/PFC enhancer were identified with the Matinspector program at http://www.genomatix.de. Sites were as follows: s1, TTTTCGGAA; s2, TTCCCCCAA; s3, TTCTTAGAA; and s4, TTTGCAGAA. Only the s3 site perfectly fits the consensus binding site (in bold) identified for D-STAT: TTCNNNGAA (34).
tsl(B-s1s2s3s4Mut)-lacZ construct.
The STAT binding sites were mutated following the protocol in the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). As a template, we used the fragment B cloned in PBSK. Once mutated, the fragment was subcloned into C4PLZ with EcoRI/SpeI. The mutagenic primers were as follows: s1Mut, TTTTCGGtt; s2Mut, TTCCCCCtt; s3Mut, TTCTTAGtt; and s4Mut, TTTGCAGtt (changed nucleotides in italics). These changes are the same that abolished expression in the even skipped stripe 3 enhancer (34).
tslGAL4 constructs.
To generate the tsl(G)GAL4 construct, an EcoRI/BamHI fragment from C4PLZ-tsl(G) was subcloned into the pPTGAL vector (35). To generate the tsl(F)GAL4 construct, an EcoRI/BamHI fragment from C4PLZ-tsl(F) was subcloned into the pChs-Gal4 vector (36).
UAS-tsldsRNA construct.
The construct was generated by using a similar strategy as the one described by Nagel et al. (37). Briefly, a BamHI/KpnI 510-bp tsl fragment was amplified by PCR with the primers cgggatccggagttctgcgagaatcg and ggggtaccgcactagccgatcgaatc (these primers were designed by using the Genome RNAi Drosophila Resources at http://www.dkfz.de/signaling2/rnai/ernai.html) and cloned into pHIBS to generate pHIBS-tsl. A SalI/KpnI fragment from pHIBS-tsl was cloned into pUAST (28) with XhoI/KpnI to generate pUAST-Hintron-tsl. Finally, an EcoRI/BamHI fragment from pHIBS-tsl was cloned into pUAST-Hintron-tsl with EcoRI/BglII.
X-Gal and Antibody Stainings.
Stainings were done by using standard protocols. All X-gal stainings were done with the same conditions: ovaries were fixed for 5 min in 2% glutaraldehyde on ice and stained overnight at 37°C. For immunostainings, the following antibodies were used: rabbit anti-β-gal at 1/1,000 (Cappel, West Chester, PA), mouse anti-FasIII at 1/20 (Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary antibodies were anti-rabbit Cy2 and anti-mouse Cy3 at 1/300 (Jackson ImmunoResearch, West Grove, PA). Nomarsky photographs were taken in a Nikon (Melville, NY) Eclipse 80i microsocope with a Nikon digital camera DXM 1200F. Confocal images were obtained with a Leica (Nussloch, Germany) SPE.
Embryonic Cuticle Preparations.
Twenty-four- to 48-h-old embryos were collected with 0.1% Triton, dechorionated with bleach, washed with 0.1% Triton, devitelinized with heptan:methanol (1:1), washed with methanol, washed with 0.1% Triton, mounted with Hoyer:lactic (1:1), and incubated at 50–60°C overnight (38). Dark-field or phase contrast photographs were taken in a Zeiss (Oberkochen, Germany) Axioskop microsocope.
Acknowledgments
We thank C. Berg (University of Washington, Seattle, WA), J. Castelli-Gair [Centro Andaluz de Biologia del Desarrollo (CABD), Seville, Spain], A. González-Reyes (CABD, Seville, Spain), S. González-Crespo (Institut de Biologia Molecular de Barcelona, Barcelona, Spain), D. Montell (Johns Hopkins School of Medicine, Baltimore, MD), L. Stevens (Albert Einstein College of Medicine, New York, NY), M. Suzanne (University of Nice, Nice, France), and the Bloomington Stock Center for providing flies and materials; S. Araujo, A. Casalí, A. González-Reyes, L. Gervais, M. Llimargas, M. Milán, and in particular D. Shaye for critically reading the manuscript; N. Martín and C. Costa for technical assistance; and J. Gallego and A. Olza for help with injections. This work was supported by grants from the Generalitat de Catalunya and the Spanish Ministerio de Educación y Ciencia. M.F. was supported by a Ramon y Cajal contract. G.V. was supported by a fellowship from Spanish Ministerio de Educación y Ciencia.
Abbreviations
- Tor
Torso
- tsl
torso-like
- PFC
posterior follicle cell
- BC
border cell
- CC
centripetal cell
- upd
unpaired
- dome
domeless.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Furriols M, Casanova J. EMBO J. 2003;22:1947–1952. doi: 10.1093/emboj/cdg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savant-Bhonsale S, Montell DJ. Genes Dev. 1993;7:2548–2555. doi: 10.1101/gad.7.12b.2548. [DOI] [PubMed] [Google Scholar]
- 3.Martin JR, Raibaud A, Ollo R. Nature. 1994;367:741–745. doi: 10.1038/367741a0. [DOI] [PubMed] [Google Scholar]
- 4.King RC. Ovarian Development in Drosophila melanogaster. New York: Academic; 1970. [Google Scholar]
- 5.Spradling AC. In: The Development of Drosophila melanogaster. Bate M, Martínez Arias A, editors. Woodbury, NY: Cold Spring Harbor Lab Press; 1993. pp. 1–70. [Google Scholar]
- 6.McGregor JR, Xi R, Harrison DA. Development (Cambridge, UK) 2002;129:705–717. doi: 10.1242/dev.129.3.705. [DOI] [PubMed] [Google Scholar]
- 7.Xi R, McGregor JR, Harrison DA. Dev Cell. 2003;4:167–177. doi: 10.1016/s1534-5807(02)00412-4. [DOI] [PubMed] [Google Scholar]
- 8.Montell DJ, Rørth P, Spradling C. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- 9.Edwards KA, Kiehart DP. Development (Cambridge, UK) 1996;122:1499–1511. doi: 10.1242/dev.122.5.1499. [DOI] [PubMed] [Google Scholar]
- 10.Niewiadomska P, Godt D, Tespass U. J Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gónzalez-Reyes A, St. Johnston D. Science. 1994;266:639–642. doi: 10.1126/science.7939717. [DOI] [PubMed] [Google Scholar]
- 12.Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, et al. Development (Cambridge, UK) 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 13.Brown S, Hu N, Castelli-Gair Hombría J. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 14.Silver DL, Montell DJ. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 15.Geisbrecht ER, Montell DJ. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Beccari S, Teixeira L, Rørth P. Mech Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- 17.Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medioni C, Cerezo D, Noselli S. Development (Cambridge, UK) 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- 18.Xu T, Caron LA, Fehon R, Artavanis-Tsakonas S. Development (Cambridge, UK) 1992;115:913–922. doi: 10.1242/dev.115.4.913. [DOI] [PubMed] [Google Scholar]
- 19.Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart M. Development (Cambridge, UK) 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 20.López-Schier H, St. Johnston D. Genes Dev. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. Curr Biol. 2003;13:1058–1063. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- 22.Rørth P. Science. 1994;266:1878–1881. doi: 10.1126/science.7997882. [DOI] [PubMed] [Google Scholar]
- 23.Borghese L, Fletcher G, Mathieu J, Atzberger A, Eades WC, Cagan RL, Rørth P. Dev Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Bo J, Bridges T, Dugan KD, Pan T, Chodosh LA, Montell DJ. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Denef N, Schüpbach T. Curr Biol. 2003;13:R388–R390. doi: 10.1016/s0960-9822(03)00317-8. [DOI] [PubMed] [Google Scholar]
- 26.Büning J. Dev Genes Evol. 2005;215:597–607. doi: 10.1007/s00427-005-0017-8. [DOI] [PubMed] [Google Scholar]
- 27.Schoppmeier M, Schröder R. Curr Biol. 2005;15:2131–2136. doi: 10.1016/j.cub.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 28.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philip AV, Yang M, Glover D, Kaiser K, et al. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 30.Ward EJ, Thaipisuttikul I, Terayama M, French RL, Jackson SM, Cosand KA, Tobler KJ, Dorman JB, Berg CA. Genesis. 2002;34:46–50. doi: 10.1002/gene.10138. [DOI] [PubMed] [Google Scholar]
- 31.Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu C, Li M, Van Den Engh G, Ruohola-Baker H. Proc Natl Acad Sci USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin GM, Spradling AC. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 33.Wharton KA, Crews ST. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- 34.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 35.Sharma Y, Cheung U, Larsen EW, Eberl DF. Genesis. 2002;34:115–118. doi: 10.1002/gene.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apitz H. Drosoph Inf Serv. 2002;85:118–120. [Google Scholar]
- 37.Nagel AC, Maier D, Preiss A. Dev Genes Evol. 2002;212:93–98. doi: 10.1007/s00427-002-0210-y. [DOI] [PubMed] [Google Scholar]
- 38.van der Meer JM. Drosoph Inf Serv. 1977;52:160. [Google Scholar]
- 39.Silver DL, Geisbrecht ER, Montell DJ. Development (Cambridge, UK) 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]