Figure 2.
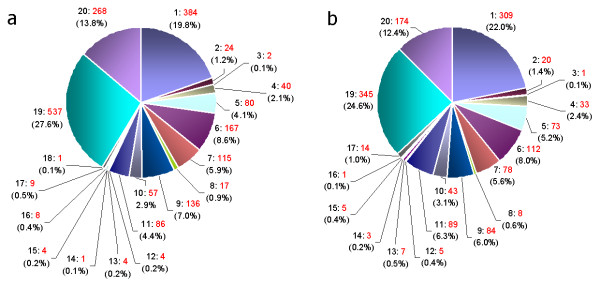
Distribution of the putative functions of 3348 ESTs from the E (a) and NE (b) library. Classification was performed according to the Munich Information centre for Proteins Sequences (MIPS) functional catalogue using BlastX search results against Arabidopsis coding sequences from the Arabidopsis genomic database (AtGDB). 1: Metabolism; 2: Energy; 3: Storage protein; 4: Cell cycle and DNA processing; 5: Transcription; 6: Protein synthesis; 7: Protein fate (folding, modification, destination); 8: Protein with binding function or cofactor requirement (structural or catalytic); 9: Cellular transport, transport facilitation and transport routes; 10: Cellular communication/signal transduction mechanism; 11: Cell rescue, defense and virulence; 12: Interaction with the cellular environment; 13: Interaction with the environment (Systemic); 14: Transposable elements, viral and plasmid proteins; 15: Cell fate; 16: Development (Systemic); 17: Biogenesis of cellular components; 18: Cell type differentiation; 19: Function unknown (regrouping Subcellular localization, Classification not yet clear cut, and Unclassified proteins); 20: No hits found.
