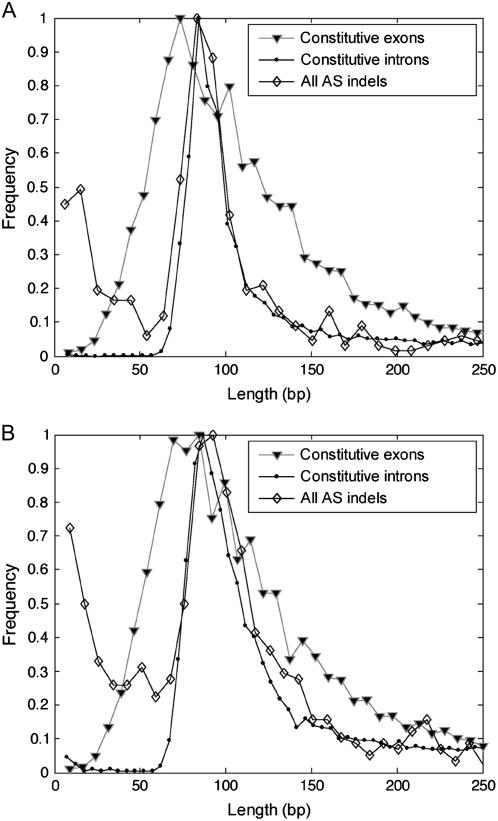
Figure 2.
Distribution of indels detected by EPGA and the distribution of constitutive introns and exons in the genomes of Arabidopsis and rice. A, Distribution of constitutive exons, constitutive introns, and indel sizes in Arabidopsis. The ESTs were aligned to the genome and constitutive exon and constitutive intron sizes were calculated from the alignment results. Constitutive introns were extracted from the genomes in the places were the ESTs had gaps in the alignment to the genome and contained the canonical borders (GT-AG, GC-AG, AT-AC). Constitutive exons were alignments to the genome situated between two constitutive introns. Indel sizes were calculated from EPGA results. B, Distribution as in A but for rice (‘japonica’ group).