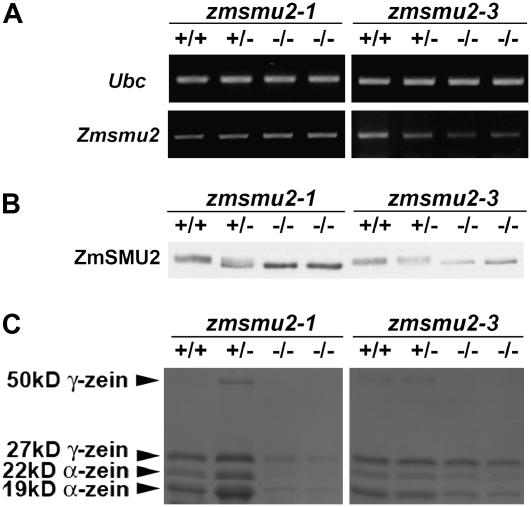
Figure 6.
zmsmu2-1 and zmsmu2-3 mutants share similar molecular phenotypes. Heterozygous (zmsmu2-1/+ or zmsmu2-3/+) plants were self-pollinated and 16-DAP kernels dissected into embryo and endosperm tissues. Embryo DNA was used to genotype the Zmsmu2 locus, and protein and RNA were extracted from homozygous wild-type, heterozygous, and homozygous mutant endosperms. +/+, Endosperm with a homozygous wild-type embryo; ±, endosperm with a heterozygous embryo; −/−, endosperm with a homozygous zmsmu2-1 mutant embryo. RNA was used for semiquantitative RT-PCR (A). Protein was separated into zein and nonzein fractions, separated by SDS-PAGE, and immunoblotted with ZmSMU2 antibodies (B, nonzeins) or stained with Coomassie Blue (C, zeins). A minimum of three replicated experiments for each homozygous zmsmu2-1 and zmsmu2-3 genotype was analyzed. A, Comparison of Zmsmu2 transcripts in F2 kernels segregating for zmsmu2-1 and zmsmu2-3. Relative to wild type, Zmsmu2 transcripts were more reduced in zmsmu2-3. Primers F5 and S1 were used for the amplification of Zmsmu2 transcript. B, Immunodetection of ZmSMU2 protein in zmsmu2-1 and zmsmu2-3. Although this protein was synthesized in both zmsmu2-3 and zmsmu2-1, its migration was altered during SDS-PAGE. C, Comparison of zein proteins in zmsmu2-1 and zmsmu2-3 endosperm. Both zmsmu2-1 and zmsmu2-3 showed reduced synthesis of zein proteins, particularly α-zeins. Zein protein designations are shown on the right; each lane contained 1/120 of the total zeins extracted from 50 mg of 16-DAP endosperm.