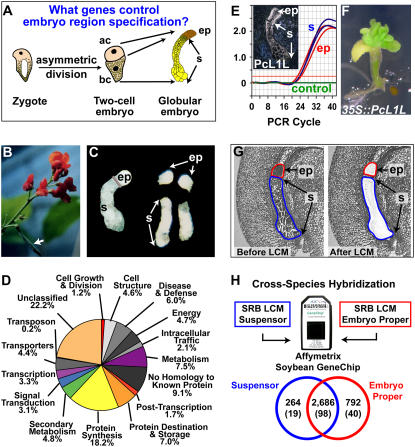
Figure 4.
Using SRB as a genomics engine to uncover genes active early in embryogenesis. A, Model for the specification of the embryo proper and suspensor, adapted and modified from Weterings et al. (2001). ac, Apical cell; bc, basal cell; ep, embryo proper; s, suspensor. B, SRB plant with a pod indicated by the arrow. C, Hand-dissected SRB globular-stage embryos before and after separating the embryo proper and suspensor. D, Functional category distribution of SRB suspensor ESTs. E, Real-time qRT-PCR validation of PcL1L mRNA accumulation pattern. Inset, In situ hybridization of SRB globular-stage embryo mRNA using a PcL1L antisense probe. In situ data were taken from Kwong et al. (2003). F, PcL1L overexpression in transgenic Arabidopsis seedlings. G, SRB globular-stage seed paraffin sections before and after capturing the embryo proper and suspensor by LCM. H, Cross-species hybridization of SRB embryo-proper and suspensor mRNA captured in G with an Affymetrix Soybean GeneChip containing 37,593 soybean probe sets. Only SRB sequences with a high similarity to soybean EST sequences on the GeneChip will hybridize. Therefore, the proportion of heterologous SRB suspensor and embryo-proper transcripts detected on the GeneChip is lower than that detected for homologous soybean RNAs (Fig. 5C). Venn diagram of transcripts detected in the suspensor and embryo proper is shown. Numbers in parentheses refer to the number of transcription factor gene transcripts detected. Data are available at http://estdb.biology.ucla.edu/seed.