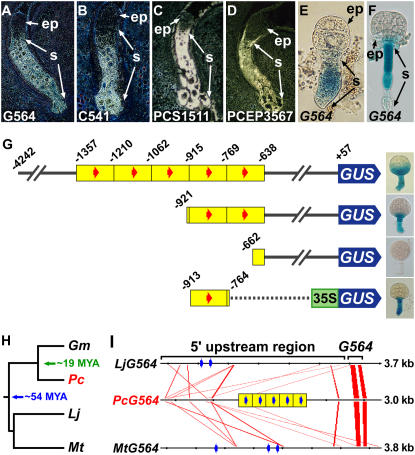
Figure 6.
Identifying DNA sequences important for suspensor transcription. A to D, In situ hybridization of SRB globular-stage embryos using antisense probes from G564 (A), C541 (B), PCS1511 (C), and PCEP3567 (D) cDNAs. G564 and C541 in situ hybridization data were taken from Weterings et al. (2001). E and F, GUS enzymatic activity in transgenic tobacco (E) and Arabidopsis (F) embryos carrying a G564∷GUS chimeric gene. G, G564 5′-deletion and gain-of-function analyses in transgenic tobacco plants. The number indicates position relative to the transcription start site (+1). Yellow blocks indicate approximately 150-bp tandem duplications. Red arrows indicate a 10-bp motif (GAAAAGC/TGAA) identified to be conserved in the upstream sequences of G564 and C541 (Weterings et al., 2001). Deletion data were taken from Weterings et al. (2001). The −913 to −764 gain-of-function construct was made by fusing this G564 upstream region with a cauliflower mosaic virus 35S∷GUS chimeric gene and transforming tobacco plants according to Koltunow et al. (1990). H, Legume phylogenetic tree, including SRB, soybean, Lotus, and Medicago. Soybean and Lotus/Medicago diverged from SRB approximately 19 and 54 million years ago, respectively (Lavin et al., 2005). I, Conserved regions among legume G564 genes. Red lines indicate closely related sequences displayed by FamilyRelationsII (20-bp window size, 75% similarity or greater; Brown et al., 2005). Blue arrows indicate motifs identified by MEME (20-bp window size; Bailey and Elkan, 1994) to be significantly enriched in the G564 upstream regions. ep, Embryo proper; Gm, G. max; Lj, L. japonicus; Mt, M. truncatula; MYA, million years ago; Pc, P. coccineus; s, suspensor.