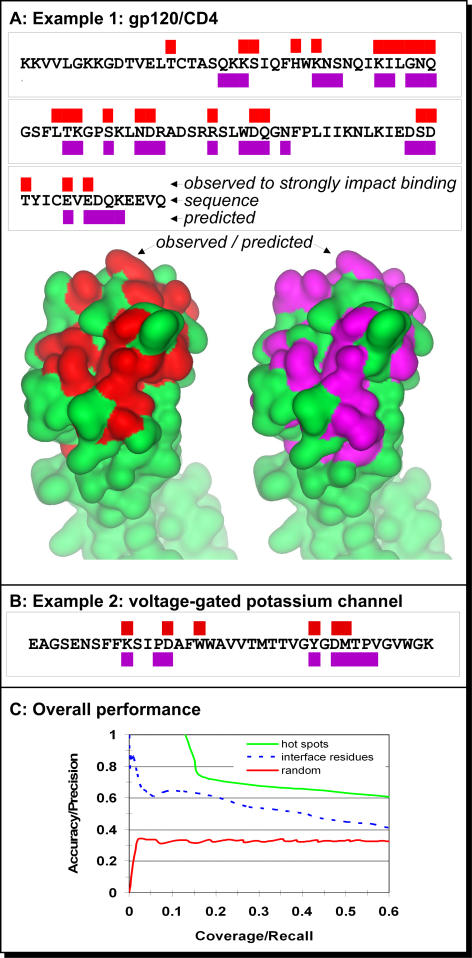
Figure 2. Accuracy of Prediction of Hotspots.
The ability to identify the residues that account for most of the energy of binding is assessed both on particular proteins and on a large dataset of alanine scans.
(A) Alanine scans and predictions of essential interface residues in the V1 domain of CD4. The red rectangles (above sequence) mark positions that were shown to have significant effect on the affinity of the binding between CD4 to gp120 upon substitution to alanine [33]; the same residues are colored in red on the lower left surface representation of CD4 (PDB ID 1wiq_A). The green rectangles (below sequence) mark positions predicted to participate in a protein–protein interaction; these residues are also colored in violet on the lower right. Note that five of the residues predicted in interfaces were not mutated in the alanine scan. Thus, we cannot evaluate their correctness and left them out of this analysis.
(B) Hotspots experimentally observed and predicted for the shaker voltage-gated potassium channel. All predictions and experimental substitutions [34] for this stretch are reported in this figure.
(C) Accuracy versus coverage in predicting hotspots and interface residues. The performance of ISIS (green) and random assignment (red) using 296 alanine scans as gold standard. The data were compiled for a set of proteins that was not used for developing the method. The stronger the confidence in our prediction, the higher the accuracy and the lower the coverage (i.e., when we select the strongest predictions [moving upward in the figure], most of these are correct). With an accuracy of approximately 0.61 (righthand side of the plot), ISIS correctly predicted most of the interacting residues in our test set. The performance of ISIS in the task for which it was originally developed, namely predicting all interface residues (broken blue), is substantially worse than its performance on hotspots.