Abstract
The crystal structure of the ferritin from the archaeon, hyperthermophile and anaerobe Pyrococcus furiosus (PfFtn) is presented. While many ferritin structures from bacteria to mammals have been reported, until now only one was available from archaea, the ferritin from Archaeoglobus fulgidus (AfFtn). The PfFtn 24-mer exhibits the 432 point-group symmetry that is characteristic of most ferritins, which suggests that the 23 symmetry found in the previously reported AfFtn is not a common feature of archaeal ferritins. Consequently, the four large pores that were found in AfFtn are not present in PfFtn. The structure has been solved by molecular replacement and refined at 2.75-Å resolution to R = 0.195 and Rfree = 0.247. The ferroxidase center of the aerobically crystallized ferritin contains one iron at site A and shows sites B and C only upon iron or zinc soaking. Electron paramagnetic resonance studies suggest this iron depletion of the native ferroxidase center to be a result of a complexation of iron by the crystallization salt. The extreme thermostability of PfFtn is compared with that of eight structurally similar ferritins and is proposed to originate mostly from the observed high number of intrasubunit hydrogen bonds. A preservation of the monomer fold, rather than the 24-mer assembly, appears to be the most important factor that protects the ferritin from inactivation by heat.
Keywords: Ferritin, Iron, Archaeon, Hyperthermophile, Ferroxidase center
Introduction
Ferritin is a protein involved in metal homeostasis by reversible storage of iron and presumably also in protection against oxidative stress by scavenging reactive oxygen species. It is a small protein of approximately 20 kDa and its main structural motif is a bundle of four parallel α-helices and a fifth short α-helix tilted towards the bundle axis and masking the one end of the cylindrical bundle. The functional ferritin molecule is an approximately 500 kDa hollow spherical assembly of 24 subunits, with outer and inner diameters of 12 and 8 nm, respectively. The 24-meric ferritin agglomerate may be a homopolymer or, in higher eukaryotes, a heteropolymer consisting of homologous subunits designated as H, M and/or L. Ferritin subunits, except the L-type subunit, incorporate a ferroxidase center (FC) that consists of a μ-oxo bridged dinuclear iron group and is responsible for iron oxidation. Additionally, a third iron site is observed in the subunits of some bacterial ferritins [1, 2]. The binuclear iron cluster of the FC and the third iron site are referred to as A, B and C iron sites, respectively. Some ferritins, also called bacterioferritins (BFRs), enclose 12 heme groups that are located between homodimeric subunit pairs, and may be involved in electron transfer [3]. The homologous protein DNA protection during starvation (DPS) and also called a “small ferritin,” is a spherical 12-subunit assembly with smaller dimensions (outer and inner diameters of 9 and 4.5 nm, respectively) and iron storage capacity (500 iron atoms vs. 3,000 in ferritin) than ferritins [4]. While iron is believed to be the main cationic substrate of ferritins, zinc accumulation in ferritin has also been reported [5].
Ferritins occur in a wide variety of organisms, from prokaryotes to mammals. In humans, their function is related to iron deficiencies or iron-overload disorders such as thalassemia, sickle cell anemia, hemochromatosis and pulmonary hemosiderosis [6–9]. Understanding the mechanism of the action of ferritin will be of great value in the treatment of iron-related diseases. Besides its medical relevance, this knowledge is also of fundamental scientific interest, especially in case of anaerobes. To date, an insight into the iron-uptake reaction has only been achieved for aerobic organisms. In these organisms, Fe(II) ions are oxidized, possibly together with phosphate oxoanions, and stored inside a ferritin shell as a soluble Fe(III) mineral, believed to be a ferrihydrite mineral with approximate composition Fe2O3·0.5H2O. Ferric ions from the core can be reduced and released from the ferritin molecule when needed by a cell. Nevertheless, the chemical nature of the physiological reductants for iron release and in the case of anaerobes that of the physiological oxidants for iron uptake have not yet been established.
Crystal structures of ferritins from bacteria, fungi, plants, insects and vertebrates are available, whereas those from archaea are limited to a ferritin from the sulfate-reducing anaerobe and thermophile Archaeoglobus fulgidus (AfFtn) (Protein Data Bank, PDB, codes 1sq3 and 1s3q) [2] and a DPS protein from Halobacterium salinarum (PDB codes 1tjo, 1tk6, 1tko and 1tkp) [10].
In AfFtn, a novel 23 point-group symmetry of the 24-mer assembly was found, which is unusual for ferritins and was only known previously in 12-meric assemblies, such as the DPS proteins [10, 11] and also in the ferritin from the bacterium Listeria innocua [12]. As a result, four outsized triangular pores about 45-Å wide are present in the protein shell of AfFtn [2].
Pyrococcus furiosus is a strict anaerobe and hyperthermophilic archaeon, living optimally at 100 °C. It was isolated from the hot marine springs off the beach of Porto di Levante in Vulcano, Italy. P. furiosus is a fermentative heterotroph that grows on starch, maltose, peptone and yeast extract and produces CO2, acetate, alanine, H2 and H2S. The expression of the native ferritin from P. furiosus (PfFtn) has been confirmed by N-terminal sequencing of the purified protein and the recombinant ferritin has been overproduced in Escherichia coli in order to afford extensive biochemical studies [13].
Herein we present the 3D structure of PfFtn, refined at 2.75-Å resolution. In contrast to the previously reported first structure of an archaeal AfFtn, the functional PfFtn 24-mer has the typical 432 point-group symmetry observed in other ferritins and BFRs. There are no four large pores such as were found in AfFtn. Instead, the characteristic threefold and fourfold channels are present in PfFtn.
The extreme thermostability of PfFtn, which in vitro can withstand 1 day of incubation at 100 °C, has been analyzed at the structural level, in comparison with structurally related ferritins. Although P. furiosus grows at the highest temperature, 100 °C, among the organisms used in thermostability analysis, our results suggest that the ferritin from Thermotoga maritima (TmFtn), which has an optimal growth temperature of 80 °C, may be even more thermostable than PfFtn. The hyperthermal stability of PfFtn and other ferritins appears to result mainly from the preservation of the monomer fold rather than the 24-mer assembly, owing to a high number of intramolecular hydrogen bonds between main-chain atoms, and between main-chain and side-chain atoms.
The FC stability was analyzed by electron paramagnetic resonance (EPR). Without further experiments, the lack of FC iron sites B and C in nonsoaked PfFtn crystals obtained does not allow any conclusions supporting or contradicting the FC as a stable catalytic cofactor.
Materials and methods
Protein production and crystallization
PfFtn was obtained and purified as previously described [13]. Ferritin iron loading prior to crystallization was performed by adding a freshly and anaerobically prepared aqueous solution of 20 mM iron sulfate, containing 0.1% HCl, to a solution of apoferritin in 50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (Hepes), pH 7, under aerobic conditions. Crystals of PfFtn were produced using the hanging drop vapor diffusion method at room temperature [14]. The crystals used for data collection were grown against 0.5 ml of 2 M ammonium sulfate reservoir solution, with drops containing 2 μl of 5 mg ml−1 protein solution in 50 mM Hepes, pH 7, and an equal amount of the reservoir solution. The crystals of the as-isolated ferritin, containing approximately 17 Fe per 24-mer, grew in 3 weeks and those loaded with approximately 1,000 Fe atoms per 24-mer prior to crystallization grew in 4 months. The crystals exhibited an irregular pyramidal shape, with base side and height between 150 and 250 μm. The iron content of as-isolated ferritin was determined with ferrene as previously described [14] by measuring the absorption of the iron–ferrene complex at 593 nm, ε = 35,500 M−1 cm−1.
Iron soaking of ferritin crystals was performed by transferring them to a 10–20-μl drop of a solution with composition 20 mM FeSO4, 2 mM Na2S2O4, 25% glycerol and 2 M ammonium sulfate, for 15 min. Zinc soaking with ZnCl2 was carried out in a similar fashion, with the exception that dithionite (Na2S2O4) was omitted from the solution.
X-ray data collection
Cryoprotecting conditions consisted of briefly dipping the crystals into a modified crystallization solution containing 20% glycerol in the case of the as-crystallized and Fe-loaded crystals, and 25% glycerol in the case of the Fe-soaked and Zn-soaked crystals, prior to flash-freezing at −173 °C in a nitrogen-gas stream, using an Oxford Cryosystems low-temperature device.
Diffraction data from an as-crystallized, an Fe-soaked and a Zn-soaked crystal of as-isolated PfFtn, as well as from a loaded PfFtn crystal, with approximately 1,000 Fe per 24-mer, were collected at the ESRF beamline BM-14, from flash-frozen single crystals at −173 °C. Two datasets were collected from the Zn-soaked crystal: the first at the Zn K-edge (9.6760 keV, 1.2810 Å, Zn peak) as determined from a fluorescence scan, and the second (9.500 keV, 1.305 Å, Zn low-E rm) on the low-energy side of the absorption edge. The diffraction images were integrated with the program MOSFLM [15]. The processed data were scaled, merged and converted to structure factors using the CCP4 program suite [16]. Data-processing statistics are summarized in Table 1.
Table 1.
X-ray diffraction data collection and processing statistics
| Beamline | ESRF BM-14 | ESRF BM-14 | ESRF BM-14 | ESRF BM-14 | ESRF BM-14 |
|---|---|---|---|---|---|
| Detector | MAR 225 CCD | MAR 225 CCD | MAR 225 CCD | MAR 225 CCD | MAR 225 CCD |
| Crystal | As-isolated | Fe-loaded | FeSO4 soak | ZnSO4 soak, Zn peak | ZnSO4 soak, Zn low-E rm |
| Wavelength (Å) | 1.7712 | 1.7340 | 1.4585 | 1.2810 | 1.305 |
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 |
| Unit cell (Å) | |||||
| a | 255.00 | 254.32 | 254.30 | 255.37 | 255.69 |
| b | 341.42 | 343.16 | 342.88 | 342.06 | 342.36 |
| c | 265.52 | 266.26 | 266.22 | 265.99 | 266.21 |
| Resolution range (Å) | 58.2–2.75 (2.90–2.75) | 63.2–2.95 (3.11–2.95) | 42.0–2.80 (2.95–2.80) | 42.9–2.80 (2.95–2.80) | 42.9–2.80 (2.95–2.80) |
| Observations | 1,685,078 (151,184) | 903,956 (85,605) | 1,011,149 (102,079) | 1,082,567 (111,068) | 1,086,719 (111,952) |
| Unique reflections | 295,335 (42,009) | 237,583 (32,389) | 280,407 (38,583) | 279,537 (37,607) | 280,474 (37,777) |
| Completeness (overall) (%) | 99.7 (97.8) | 98.1 (92.2) | 99.1 (94.1) | 98.7 (91.6) | 98.7 (91.9) |
| Redundancy | 5.7 (3.6) | 3.8 (2.6) | 3.6 (2.6) | 3.9 (3.0) | 3.9 (3.0) |
| Rmerge | 0.085 (0.300) | 0.113 (0.532) | 0.081 (0.354) | 0.075 (0.288) | 0.077 (0.344) |
| I/σ(I) | 16.5 (3.1) | 10.8 (1.6) | 8.2 (2.0) | 8.8 (2.5) | 8.6 (1.9) |
| Anomalous completeness (%) | 98.0 (88.3) | 89.2 (71.1) | 94.4 (64.8) | 97.7 (85.4) | 97.6 (85.5) |
| Anomalous redundancy | 2.9 (1.9) | 2.0 (1.5) | 1.8 (1.6) | 1.9 (1.5) | 2.0 (1.5) |
| Ranom | 0.043 (0.204) | 0.078 (0.414) | 0.058 (0.294) | 0.063 (0.252) | 0.059 (0.288) |
| Estimated Boverall (Å2) | 53.3 | 62.7 | 60.5 | 54.3 | 58.5 |
Values in parentheses refer to the last resolution shell
Structure determination of as-isolated and Fe-loaded PfFtn
Previously [14], the 3D structure of PfFtn was solved by the molecular replacement (MR) method, using the program PHASER [17] and the homologous TmFtn (PDB entry 1vlg) as the search model. The sequence of this 165 amino acid ferritin shares a 51% amino acid sequence identity (88 amino acid residues) with that of PfFtn (174 amino acids), and the only insertions or deletions occur at the termini of the subunit chains. However, despite the success of the MR calculations we believed that, because of the low resolution of the diffraction data, relatively low amino acid sequence identity with the search model, as well as the complexity of the structure, experimental phase information would be needed in order to obtain the best possible structural model from the data. To that end, the first BM-14 dataset was collected at a long wavelength (7.0000 keV, 1.7712 Å) from a crystal of as-isolated PfFtn with the intention of using S-SAD in phasing, whereas a second dataset was collected just above the Fe K-absorption edge (7.1500 keV, 1.7340 Å) from an Fe-loaded PfFtn crystal, with the aim of using Fe-SAD in phasing and also for better location of the incorporated Fe atoms. However, neither dataset could provide the desired independent phase information, owing to complications that arose from radiation damage, which prevented us from collecting the highly redundant dataset required for application of the S-SAD method to the first dataset, as well as the lack of a sufficiently high ordered Fe content, which limited the usefulness of the anomalous dispersion data obtained from the second dataset.
Therefore, the 3D structure of PfFtn was solved by repeating the MR calculations previously described [14] with the new “as-isolated” 2.75-Å dataset, using the standard PHASER protocol as implemented through the CCP4 graphical user interface, with reflection data up to 3.25-Å resolution. Previously, we had established [14] that the asymmetric unit of the PfFtn crystal structure contained 36 monomers, arranged as 1.5 24-mers. The search model used was half of a TmFtn 24-mer, constructed from the PDB 1vlg coordinates without the Fe sites, and three such models were searched for in the crystal structure of PfFtn. The MR calculations with PHASER gave a single solution. Prior to model rebuilding, electron-density map improvement was carried out both by using Arp/wArp [18] in atom update and refinement mode as well as by using DM [19] with 36-fold noncrystallographic symmetry (NCS) averaging. In this way, the initial PHASER figure of merit of 0.502 for the full resolution range of the 2.75-Å dataset was improved to 0.697 (Arp/wArp) and 0.849 (DM). Rebuilding of the protein chain to take into account the differences in sequence and chain length between TmFtn and PfFtn was carried out with the program TURBO [20] using one monomer and the 36-fold averaged electron-density map obtained from the DM calculations. The coordinates for each of the remaining 35 monomers were obtained from those of the first by applying the appropriate NCS transformations, followed by model correction with TURBO using the electron-density maps obtained from both the DM and the Arp/wArp calculations. In these maps, peaks corresponding to Fe occupation of the A sites only in the FC were observed, and therefore only 36 iron sites (one per monomer), were included in the model.
The structure of the Fe-loaded PfFtn was also determined by MR with PHASER using a similar procedure; however, the |Fo| density map, as well as an anomalous electron density map, calculated with MR phases, showed that only the A site in the FC was occupied by Fe atoms. Since the “as-isolated” dataset had higher resolution and better quality, further work on the “Fe-loaded” dataset was abandoned.
Structure refinement of as-isolated PfFtn
Refinement was carried out with the program REFMAC [21] using medium main-chain and weak side-chain NCS restraints between the 18 dimers in the asymmetric unit. At a later stage, a translation–libration–screw (TLS) rigid-body motion refinement [22] was made prior to restrained refinement of atomic positions and thermal motion parameters. One rigid body was defined for each of the 36 independent monomers. In the final refinement stages, a total of 832 solvent molecules were located with Arp/wArp [23]. On the basis of their B factors, observed electron density and interactions with neighboring protein residues, 791 were assigned to water molecules, while the remaining 41 were modeled as sulfate ions. Individual restrained B factors were refined for all non-hydrogen atoms and, for consistency with the structure refinements of the Fe-soaked and Zn-soaked crystals (see below), only tight NCS restraints (0.1 Å positional and 1.0 Å2 thermal parameter) were applied to all the protein residues involved in the FC metal coordination, instead of defining the coordination geometry for each individual protein–metal bond as in the earlier stages of refinement.
The final values of R and Rfree were 0.195 and 0.247, respectively. Neither TLS rigid-body refinement nor full NCS restraints (i.e., extending over the whole monomer and relating all the chains in the asymmetric unit) were used in the final refinement, as they were seen to lead to higher values of both R and Rfree. Rfree [24] was calculated from a randomly chosen subset of the data, containing approximately 5% of the total number of independent reflections. Throughout the refinement, the model was periodically checked and corrected with TURBO against 2|Fo| − |Fc| and |Fo| − |Fc| electron-density maps. The refinement statistics are included in Table 2. The maximum-likelihood estimate of the overall coordinate error, obtained with REFMAC, was 0.20 Å. For all 36 independent monomers in the PfFtn crystal structure, only the first 168 residues were included in the model. Although some electron density was visible beyond Gly168, it was not possible to build any more C-terminal residues into it, and it is likely that the C-terminal tail is disordered in all the independent PfFtn monomers. On the basis of a B-value comparison with the protein residues directly coordinating the iron atoms, it was realized that at least 17 out of the 36 metal sites were not fully occupied, and were assigned occupation factors of 0.8 (0.7 in one case). In addition, 788 side-chain atoms could not be seen in either 2|Fo| − |Fc| or |Fo| − |Fc| electron-density maps and were therefore given zero occupancy during the refinement.
Table 2.
Refinement and structure analysis statistics for the ferritin from Pyrococcus furiosus (PfFtn)
| Crystal | As-isolated, as-crystallized | Fe-soaked | Zn-soaked |
|---|---|---|---|
| Resolution (Å) | 56.8–2.75 | 42.0–2.80 | 42.8–2.80 |
| No. of reflections in work set/test set | 280,375/14,914 | 266,173/14,187 | 265,356/14,134 |
| Rwork/Rfree | 0.195/0.247 | 0.197/0.249 | 0.201/0.251 |
| Number of atoms | |||
| Protein—total number of atoms | 49,824 | 49,824 | 49,824 |
| Protein—atoms with zero occupancy | 788 | 718 | 720 |
| Fe/Zn atoms | 36 | 108 | 108 |
| Water molecules (sulfate ions) | 791 (41) | 413 (26) | 162 (44) |
| Mean B factors (Å2) | |||
| Protein main chain (side chain) | 32.6 (34.6) | 38.0 (39.9) | 30.9 (32.7) |
| Fe/Zn atoms | 57.5 | 53.2 | 36.9 |
| Water molecules (sulfate ions) | 26.5 (67.1) | 29.0 (76.8) | 21.9 (74.0) |
| Root mean square deviations from ideal values | |||
| Bond lengths (Å) | 0.014 | 0.014 | 0.015 |
| Bond angles (°) | 1.44 | 1.46 | 1.48 |
| Mean positional error (Å) | 0.20 | 0.22 | 0.22 |
| PROCHECK Ramachandran analysis | |||
| Total no. analyzed residues | 5,544 | 5,544 | 5,544 |
| Residues in core regions, no. (%) | 5,255 (94.79) | 5,241 (94.53) | 5,239 (94.50) |
| Residues in allowed regions, no. (%) | 277 (5.00) | 285 (5.14) | 289 (5.21) |
| Residues in generous regions, no. (%) | 11 (0.20) | 10 (0.18) | 9 (0.16) |
| Residues in disallowed regions, no. (%) | 1 (0.02) | 8 (0.14) | 7 (0.13) |
| PROCHECK G factors | |||
| Mean (range) dihedral | 0.00 (−0.13 to 0.07) | −0.04 (−0.23 to 0.03) | −0.04 (−0.21 to 0.09) |
| Mean (range) covalent | 0.38 (0.32–0.45) | 0.38 (0.29–0.46) | 0.36 (0.26–0.44) |
| Mean (range) overall | 0.15 (0.07–0.19) | 0.12 (0.00–0.18) | 0.12 (0.01–0.17) |
The structure was analyzed with PROCHECK [25] and its stereochemical quality parameters were within their respective confidence intervals. A Ramachandran [26] ϕ,φ plot showed that of the 5,544 non-glycine and non-proline residues, 11 (0.20%) were found to lie in the generously allowed regions and only one (0.02%) outlier was observed. All these residues with more unusual ϕ,φ conformations were located in a loop region containing Lys145, for which the electron density was generally poor. The final atomic coordinates were deposited with the PDB [27], to be released upon publication, with the accession code 2jd6.
Structure determination and refinement of the Fe-soaked and Zn-soaked PfFtn crystals
In view of the very small changes in cell parameters observed in comparison with those of the as-isolated crystal, the structures of the Fe-soaked and Zn-soaked crystals of PfFtn were determined directly by using the final refined coordinates of the as-isolated structure, omitting the iron sites and the solvent atoms. Ten cycles of rigid-body refinement were first carried out, followed by a TLS rigid-body motion refinement [22] prior to restrained refinement of atomic positions and thermal motion parameters. At this stage, the 2|Fo| − |Fc| or |Fo| − |Fc| electron-density maps were inspected with COOT [28] for inclusion of the FC A, B and C metal sites in the models. Because of the complexity of the metal coordination in the FC, the low resolution of the diffraction data and the realization that some of the FC metal sites were not fully occupied (see “Discussion” for details), tight NCS restraints (0.1 Å positional and 1.0 Å2 thermal parameter) were applied to all the protein residues involved in the FC metal coordination, instead of defining the cordination geometry for each individual protein–metal bond.
In the final refinement of the Fe-soaked crystal structure, 423 solvent molecules and 26 sulfate ions were located with Arp/wArp [23] and included in the model, and individual restrained B factors were refined for all non-hydrogen atoms. On the basis of a B-value comparison with the protein residues directly coordinating the iron atoms, it was realized that not all metal sites were fully occupied: all the B sites and one C site were assigned an occupation of 0.5. The final values of R and Rfree were 0.197 and 0.249. The refinement statistics are listed in Table 2. The structure was analyzed with PROCHECK [25] and its stereochemical quality parameters were within their respective confidence intervals. A Ramachandran [26] ϕ,φ plot showed that of the 5,544 non-glycine and non-proline residues, ten (0.18%) were found to lie in the generously allowed regions and only eight (0.14%) outliers were observed. With three exceptions, all these residues with more unusual ϕ,φ conformations were located in a loop region containing Lys145, for which the electron density was generally poor.
In the final refinement of the Zn-soaked crystal structure, 162 solvent molecules and 44 sulfate ions were located with Arp/wArp [23] and included in the model, and individual restrained B factors were refined for all non-hydrogen atoms. A B-value comparison with the protein residues directly coordinating the metal atoms, combined with the observation of a mixed iron/zinc population at the A sites (see below for details) led us to postulate a mixed (50:50 Zn/Fe) occupancy for the A sites, which was modeled as a Zn atom with 0.9 occupancy. Also, the B sites were modeled as partially occupied by zinc (50%) and the C sites as partially occupied by iron (50%). The final values of R and Rfree were 0.201 and 0.251. The refinement statistics are included in Table 2. The structure was analyzed with PROCHECK [25] and its stereochemical quality parameters were within their respective confidence intervals. A Ramachandran [26] ϕ,φ plot showed that of the 5,544 non-glycine and non-proline residues, nine (0.16%) were found to lie in the generously allowed regions and only seven (0.13%) outliers were observed. With two exceptions, all these residues with more unusual ϕ,φ conformations were located in a loop region containing Lys145, for which the electron density was generally poor.
As for the as-isolated structure, neither TLS rigid-body refinement nor full NCS restraints were used in the final refinement of the Fe-soaked or Zn-soaked structures, as they were seen to lead to higher values of both R and Rfree. Rfree [24] was calculated from a randomly chosen subset of the data, containing approximately 5% of the total number of independent reflections, and with the same h,k,l indices as for the “as-isolated” data. These test-set reflections were not chosen in thin resolution shells. However, while it is true that the 24-mers have very high NCS, in all three refined crystal structures the Rfree/Rwork ratios are typical for the data resolution and refinement parameters used, [29] and therefore there is no indication that the more simplistic test-set choice may have introduced a bias in the Rfree values. Throughout the refinement, the models were periodically checked and corrected with COOT [28] against 2|Fo| − |Fc| and |Fo| − |Fc| electron-density maps. Since the asymmetric unit of both structures contains 36 monomers, and each monomer contains one FC with three metal sites, a total of 108 metal atoms are present in the model. In addition, 718 side-chain atoms could not be seen in either 2|Fo| − |Fc| or |Fo| − |Fc| electron-density maps for the Fe-soaked structure (720 for the Zn-soaked structure) and were given zero occupancy during the refinement. The final atomic coordinates were deposited with the PDB [27], to be released upon publication, with the accession codes 2jd7 (Fe-soaked structure) and 2jd8 (Zn-soaked structure).
Results and discussion
Structure of the PfFtn monomer
The PfFtn monomer exhibits the typical four-helical bundle fold which is encountered in ferritins, BFRs, DPS proteins and rubrerythrins. According to the DSSP algorithm [30] the four helices comprise residues 4–34 (helix A), 37–64 (helix B), 83–111 (helix C) and 114–144 (helix D). A short, C-terminal α-helix (helix E, residues 148–160), bordering one of the cylindrical bundle openings, followed by a tail without any secondary structure also occurs in the PfFtn monomer fold (Fig. 1a, b). For convenience of terminology, the two extremities of the cylindrical α-helical bundle will be further referred to as the open side and the E-helix sides of the monomer.
Fig. 1.
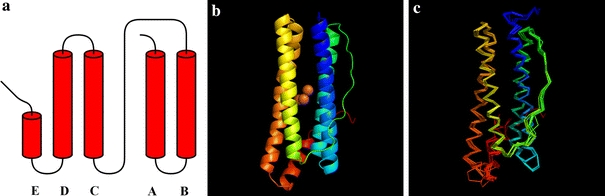
a Topology diagram of the ferritin from Pyrococcus furiosus (PfFtn). b Ribbon diagram of PfFtn showing the location of the ferroxidase center sites A, B and C (orange spheres) near the center of the four-helical bundle. c Superposition of the PfFtn Cα chain with those of its eight closest homologous structures. In b and c, the molecule colors change from blue to red in the N-terminal to C-terminal direction. b, c Prepared with PyMOL [50]
The PfFtn monomer is built of hydrophobic amino acids at its open and E-helix sides and polar and hydrophilic residues in the middle of the subunit, similarly to the subunits from other ferritins. The open side of the monomer contains three leucine residues (Leu10, Leu104 and Leu120), two phenylalanine residues (Phe57 and Phe123), Met6, Tyr60 and Ile101, whereas the E-helix side has three phenylalanine residues (Phe87, Phe31 and Phe39), Trp42 and Met27 (denoted by asterisks in Fig. 2). The residues defining the open and E-helix sides of the monomer are conserved among PfFtn, the ferritin from E. coli (EcFtnA), AfFtn and human H chain ferritin (HuHF) in terms of their polarities, apart from Phe123, which in other ferritins is a tyrosine.
Fig. 2.
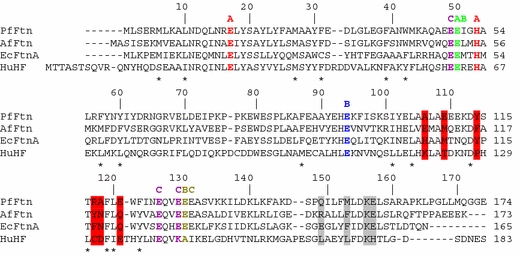
Amino acid sequence alignment of PfFtn, the ferritin from Archaeoglobus fulgidus (AfFtn), the ferritin from Escherichia coli (EcFtnA) and human H chain ferritin (HuHF). Asterisks show the residues around the open and E-helix sides of the monomer; A, B and C denote the residues coordinating the iron sites of the ferroxidase center (FC); red bars highlight the main residues located around the threefold channels and gray bars those around the fourfold channels. The alignment was prepared with ClustalW [51]
The central part of the subunit comprises the negatively charged and polar amino acids involved in metal binding. These are Glu17, Glu49, Glu50, Glu94, Glu126, Glu129 and Glu130 and His53, which are shown in Fig. 2 with A, B or C labels to denote the iron site they coordinate (see “The FC”). In addition, Tyr24 and Gln127, which in EcFtnA [1] indirectly contribute to the A and B sites via hydrogen bonds to Glu94 and Glu17, are also oriented towards the FC in PfFtn.
It has been noted [2] that Gly67, Ser84 and Gly147 (in AfFtn numbering) are conserved among AfFtn, EcFtnA and HuHF, and define the BC loop and the end of D helix, respectively. In the case of PfFtn, only Ser84 is conserved, while the two glycine residues are changed into Asn65 and Lys145.
A surface hydrophobic pocket at the PfFtn dimer interface
A large surface pocket, largely lined by hydrophobic residues, was located at the PfFtn dimer interface. Interestingly, one of these is Ile51, corresponding to Met 57 in the BFR from Desulfovibrio desulfuricans ATCC 27774 [31], which binds the heme group. Instead of a heme, the pocket appears to be occupied by a disordered string of water molecules (Fig. 3) which form a continuous region of electron density. This feature was observed for all the PfFtn structures investigated. A similar situation was reported by Yu et al. [32] for the hydrophobic cavity of human interleukin 1β.
Fig. 3.
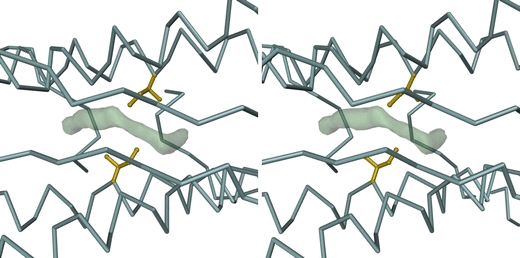
Stereoview of a dimer interface in the as-isolated PfFtn structure, showing a difference electron density feature, drawn at the 2.5 map root mean square (RMS) level, which results from a disordered chain of water molecules trapped within a hydrophobic pocket centered around Ile51. This feature is observed in all dimers of all PfFtn structures investigated. The bulk of the PfFtn dimer is represented by its Cα trace (blue-gray). The side chains (including Cα atoms) of the Ile51 residues in both monomers are represented in ball-and-stick mode and are colored gold. The view is down the noncrystallographic twofold symmetry axis of the dimer, looking towards the inside of the PfFtn 24-mer. The figure was prepared with DINO [52]
Subunit structure comparison
In order to compare the PfFtn structure with the structures of other known ferritins, a protein structure comparison against the PDB using the secondary structure matching (SSM) tool at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/ssm) [33] was performed. This resulted in 377 hits, which, after elimination of duplicate protein chains, were seen to correspond to 63 different protein structures. A multiple structural alignment was then carried out with a subset of eight SSM hits, listed in Table 3 and shown as a superposition of Cα chains in Fig. 1c. The members of this subset were chosen such that the percentage of their aligned residues was equal to or greater than 25%, all five secondary structure elements (α-helices) were aligned, the Q score, of their pairwise SSM alignment with PfFtn was greater than 0.5, and considering only one crystal form of each different protein structure from each organism. This subset includes representatives from archaea, bacteria and eukarya. The structural alignment showed PfFtn to be highly similar to TmFtn, followed by AfFtn and EcFtnA (Table 3). The deviations occur mostly in loop regions: AB loop from Glu32 to Leu34, end of the BC loop from Lys78 to Pro83, CD loop and helix D up to Phe123 and DE loop from Lys145 to Leu151 (in PfFtn numbering).
Table 3.
Proteins with known 3D structure most closely homologous to PfFtn
| PDB ID | Protein name | Source organism | Chain length | Identity (%) | Identity SS-aligned (%) | Q scorea | RMSD (Å) |
|---|---|---|---|---|---|---|---|
| 1vlg | Ferritin (TM1128) | Thermotoga maritima | 176 | 55 | 55.3 | 0.92 | 0.49 |
| 1s3q | Ferritin | Archaeoglobus fulgidus | 173 | 50 | 48.2 | 0.94 | 0.53 |
| 1krq | Ferritin | Campylobacter jejuni | 167 | 39 | 39.2 | 0.81 | 1.02 |
| 1eum | Ferritin ECFTNA | Escherichia coli | 165 | 38 | 38.4 | 0.86 | 0.90 |
| 1r03 | Mitochondrial ferritin | Homo sapiens | 182 | 40 | 30.6 | 0.68 | 1.55 |
| 2fha | H ferritin K86Q mutant | Homo sapiens | 183 | 37 | 29.4 | 0.62 | 1.98 |
| 1mfr | M ferritin | Rana catesbeiana | 176 | 36 | 29.6 | 0.68 | 1.67 |
| 1rcg | L ferritin | Rana catesbeiana | 173 | 32 | 25.0 | 0.61 | 2.03 |
PDB Protein Data Bank, SS secondary structure, RMSD root mean square deviation
aParameter that takes the alignment length and the RMSD between the superimposed atoms into account, and varies from 0 in the case of poor matches to 1 in the case of identical proteins
Structure of the PfFtn 24-mer and comparison with AfFtn
A highly conserved quaternary structure appears among all known ferritins—a spherical tetraeicosameric assembly with 432 point-group symmetry. An interesting exception to this rule is the recently reported structure of the AfFtn 24-mer [2], which exhibited 23 point-group symmetry, normally an attribute of the dodecameric DPS proteins. Even though PfFtn and AfFtn are highly similar archaeal ferritins (50 and 70% amino acid sequence identity and similarity, respectively), the PfFtn 24-mer has the canonical 432 point-group symmetry instead of the 23 point-group symmetry found in the AfFtn 24-mer. Although there may be other archaeal tetraeicosameric ferritins with 23 point-group symmetry like AfFtn, our structure clearly suggests that it is not a feature of all archaeal ferritins.
When comparing the two quaternary structures (Fig. 4), it can be seen that the 432 and 23 arrangements in PfFtn and AfFtn result from a different organization of the four fundamental units, hexamers formed by three dimers around a 90° vertical turn from a threefold NCS rotation axis. The 23 arrangement leads to the appearance of four outsized triangular pores about 45-Å wide in the protein shell of AfFtn [2], which are absent in the 432 arrangement of PfFtn and all other known structures of tetraeicosameric bacterial ferritins.
Fig. 4.

Ribbon diagrams of the PfFtn and AfFtn 24-mers. I View down a threefold axis, showing one “fundamental hexamer.” II View rotated by approximately 90° about a vertical axis, showing the different hexamer arrangement, which leads to the appearance of large pores in AfFtn. The figure was prepared with PyMOL [50]
Threefold and fourfold channels
The eight threefold channels are formed by helices C and D from three adjacent dimers, in such a way that, with respect to the hollow tetraeicosameric protein shell, the C-terminal ends of helices C define the outer entrance to the channel and the N-termini of helices D define the inner entrance (Fig. 5). The PfFtn threefold channel is lined by a mix of hydrophobic and hydrophilic amino acids Ala106 and Glu109 at the outer side of the channel, Tyr114 and Arg117 in the middle and Ala118 and Glu121 at the inner entrance of the channel (Fig. 5). In general, and similarly to EcFtnA and AfFtn, this channel is less hydrophilic than in vertebrate H and L chain ferritins, suggesting a rather hydrophobic threefold channel in microbial ferritins in comparison with vertebrate ferritins. Nevertheless, when compared with that in EcFtnA and AfFtn, the threefold channel in PfFtn is more similar to that of HuHF, in the sense that is it more negatively charged at its inner entrance and more positively charged in its central region (Fig. 5).
Fig. 5.
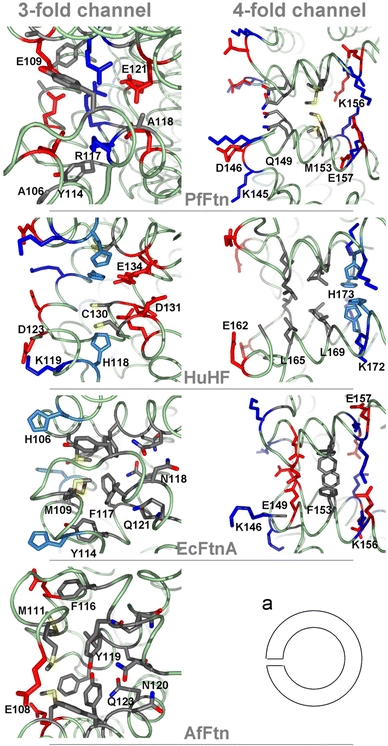
Profile view of the threefold and fourfold channels in PfFtn, AfFtn, EcFtnA and HuHF. The exterior of the shell lies on the left side and the inner cavity on the right side of each cartoon as shown schematically in a. The red residues correspond to acidic Glu and Asp residues and the blue ones to positive Lys, Arg and His residues, and highlight the arrangement of positive and negative amino acids along the channels. The figure was prepared with PyMOL [50]
On the basis of the crystallographic, kinetic, mutagenesis and electrostatic calculations studies on HuHF [9, 34–36], the threefold channels were proposed to be involved in the entry of iron ions into the ferritin molecule. The inhibition of iron uptake in HuHF upon Asp131 and Glu134 substitutions [34] and the coordination of Ca2+ by Asp131 and Glu134 [37] have been observed in this protein. The inner narrow entrance of the threefold channel in PfFtn resembles that of HuHF, suggesting that a similar mechanism is possible in PfFtn. Nevertheless, the mutations of His118 and Cys130 also influenced the iron-binding capacity of HuHF and inhibited the binding of different metals by the channel to different extents. The different amino acid arrangements in the threefold channel of the various ferritins suggests the possibility of different iron incorporation mechanisms.
The six fourfold channels, formed around the fourfold symmetry axes of the PfFtn 24-mer, are lined, from the outside to the inside of the protein shell, by Gln149, Met153, Lys156 and Glu157 (Fig. 5). Owing to a nozzlelike shape formed by Gln149, the PfFtn fourfold channel is longer (approximately 16 Å) than its counterparts in EcFtnA (approximately 14 Å) and HuHF (approximately 12 Å). In PfFtn, Lys145 in the DE loop, which forms an outer gate to the fourfold channel, occurs in place of EcFtnA Gly145 or HuHF Gly159, making this gate more polar and charged in PfFtn. In addition, the DE loop and its amino acids are somewhat oriented outwards from the fourfold channel, resulting in a hourglass shape of this channel, which is less strongly developed in EcFtnA and even less so in HuHF, where the pore has an almost cylindrical shape. The outer entrance of the PfFtn fourfold channel is polar and hydrophilic, the region inside the channel is nonpolar and the inner entrance is polar again. The fourfold channels in EcFtnA have a similar arrangement of polar residues at both channel entrances and are nonpolar in the middle, while in HuHF they are mostly apolar with Leu165 and Leu169 and polar His173 on the inner side of the channel (Fig. 5). In electrostatic terms, the PfFtn fourfold channel is similar to that of both EcFtnA and HuHF; the outer entrance of it is uncharged as in HuHF and the inner entrance has a compensated charge from the sequence of positive Lys156 and negative Glu157 as in EcFtnA.
The FC
The active site responsible for the oxidation of Fe(II) to Fe(III) in ferritins, the FC, is located in the central part of each subunit (Fig. 1b). It is generally believed that in most ferritins the initial stages of the core formation take place via the FC. Subsequently, iron incorporation and removal is thought to occur predominantly through one or the other of the channels formed around the threefold and fourfold symmetry axes of the 24-mer. In this discussion, the nomenclature for the FCs in ferritins and BFRs presented in a recent review [38] will be followed. The FC in PfFtn is of the FtnA (EcFtnA) type, which is strictly conserved in all the closest prokaryote structural homologues of PfFtn besides EcFtnA (Table 4). The EcFtnA FC contains three metal binding sites (A, B and C), in contrast with the HuHF and E. coli BFR FC, which only contain two (A and B). The FC of the as-crystallized PfFtn showed iron bound to site A only. Iron binding to sites B and C was observed only after soaking the PfFtn crystals in Fe(II).
Table 4.
The ferroxidase center in PfFtn and its homologous crystallized ferritins
| PDB ID | Protein name | Source organism | Resolution (Å) | FC type | Metal site occupation |
|---|---|---|---|---|---|
| – | Ferritin | Pyrococcus furiosus | 2.75 | FtnA | A (Fe) A, B, C (Fe,Zn) |
| 1s3q | Ferritin | Archaeoglobus fulgidus | 2.10 | FtnA | A, B (Zn)a A, B, C (Fe) |
| 1vlg | Ferritin (TM1128) | Thermotoga maritima | 2.00 | FtnA | A (Fe) |
| 1krq | Ferritin | Campylobacter jejuni | 2.70 | FtnA | A, B (Ow?)b |
| 1eum | Ferritin ECFTNA | Escherichia coli | 2.05 | FtnA | A, B (Zn)c A, B, C (Fe) |
| 1r03 | Mitochondrial ferritin | Homo sapiens sapiens | 1.70 | HuHF | – |
| 2fha | H ferritin K86Q mutant | Homo sapiens sapiens | 1.90 | HuHF | – |
| 1mfr | M ferritin | Rana catesbeiana | 2.80 | HuHF | – |
| 1rcg | L ferritin | Rana catesbeiana | 2.20 | – | – |
FC ferroxidase center
aZn in 1s3q and Fe in 1sq3 [2]
b1krq contains four water molecules, two of which are located at the FC, in positions corresponding to A and B sites. They might be low-occupancy metal sites but there is no indication of whether it is a native or a soaked crystal
c1eum corresponds to the native structure [1]; these sites were located in the Zn2+ and Fe3+ derivatives, respectively, for which no coordinates were deposited in the PDB
In PfFtn, iron site A is coordinated by Glu17, Gly50 and His53, iron site B by Glu50, Glu94 and Glu130, and iron site C by Glu49, Glu126, Glu129 and Glu130. Site B is 3.0 Å distant from site A. Site C is located 7.5 and 6.3 Å away from sites A and B, respectively (Table 5, Fig. 6). Glu17 and Glu94 appear to be monodentate ligands; therefore, sites A and B are coordinated by side chains from three amino acids, while site C is coordinated by side chains from four amino acids. The final 2|Fo| − |Fc| electron density around the FC of a selected PfFtn monomer in the as-isolated, Fe-soaked and Zn-soaked crystal structures are represented in Fig. 7a–c, respectively.
Table 5.
Coordination geometry statistics of the ferroxidase center in PfFtn
| Bondsa | Distance (Å) | ||
|---|---|---|---|
| As-isolatedb | Fe-soakedb | Zn-soakedb | |
| MA...Glu17 Oε1 | 2.24 (0.09) [2.00–2.46] | 2.21 (0.08) [2.00–2.47] | 2.18 (0.07) [2.01–2.32] |
| MA...Glu50 Oε1 | 2.26 (0.11) [2.09–2.47] | 2.24 (0.09) [2.09–2.55] | 2.22 (0.07) [2.02–2.38] |
| MA...His53 Nδ1 | 2.12 (0.09) [1.97–2.41] | 2.28 (0.09) [2.12 – 2.47] | 2.21 (0.08) [2.08 – 2.40] |
| MA...MB | 3.02 (0.19) [2.62–3.63] | 2.93 (0.18) [2.54–3.49] | |
| MA ... MC | 7.41 (0.10) [7.18–7.63] | 7.51 (0.11) [7.30–7.77] | |
| MB...Glu50 Oε2 | 2.21 (0.12) [1.93–2.45] | 2.16 (0.12) [1.92–2.50] | |
| MB...Glu94 Oε2 | 2.34 (0.14) [2.12 – 2.70] | 2.40 (0.10) [2.18 – 2.65] | |
| MB...Glu130 Oε1 | 2.87 (0.16) [2.47–3.14] | 2.81 (0.20) [2.41–3.21] | |
| MB...MC | 6.29 (0.20) [5.81–6.77] | 6.25 (0.15) [5.93–6.55] | |
| MC...Glu49 Oε1 | 2.11 (0.08) [1.92 – 2.36] | 2.29 (0.11) [2.13 – 2.56] | |
| MC...Glu126 Oε1 | 2.06 (0.09) [1.87–2.26] | 2.31 (0.16) [1.84–2.53] | |
| MC...Glu129 Oε1 | 2.31 (0.10) [2.12–2.58] | 2.02 (0.10) [1.81–2.26] | |
| MC...Glu130 Oε2 | 1.89 (0.08) [1.72–2.04] | 1.89 (0.09) [1.66–2.07] | |
aM stands for metal (Fe in as-isolated and in Fe-soaked; Zn or Fe in Zn-soaked)
bFor each column, the numbers listed are the mean value, calculated from the 36 independent distances in the asymmetric unit, followed by the corresponding standard deviation in parentheses, and finally the minimum and maximum values in the population, enclosed in brackets
Fig. 6.
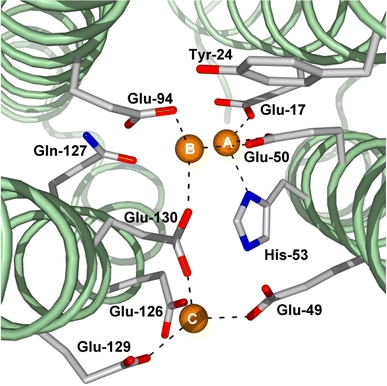
View of the FC down the PfFtn subunit from the E-helix side. The three metal sites are annotated as A, B and C. Metal occupation of sites B and C is observed only upon crystal soaking in Zn or Fe. The dashed lines correspond to the coordination geometries described in Table 5. The figure was prepared with PyMOL [50]
Fig. 7.
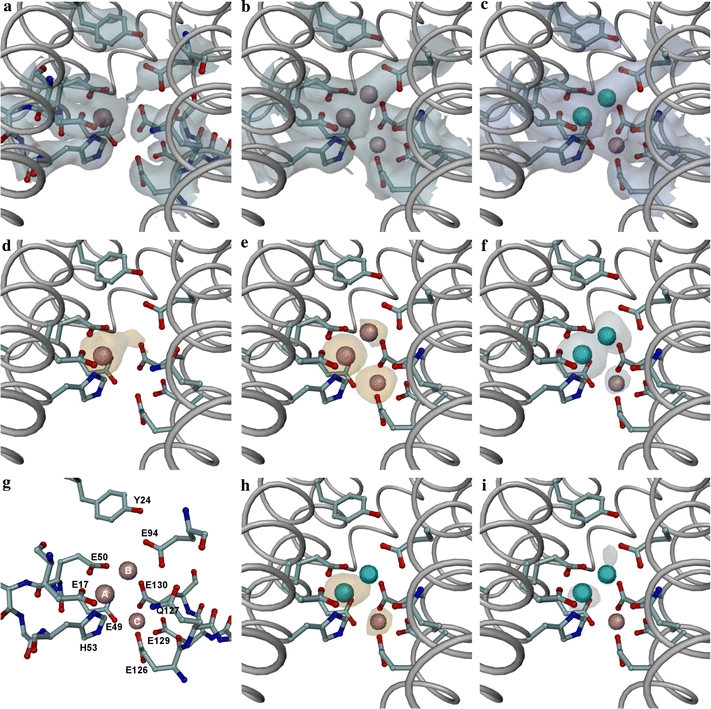
Details of the FC of a selected monomer in the structures of as-isolated, Fe-loaded, Fe-soaked and Zn-soaked PfFtn crystals. a Final 2|Fo| − |Fc| electron-density map for the as-isolated PfFtn, contoured at 1.2 map RMS. b Final 2|Fo| − |Fc| electron-density map for the Fe-soaked PfFtn, contoured at 1.2 map RMS. c Final 2|Fo| − |Fc| electron-density map for the Zn-soaked PfFtn, contoured at 1.2 map RMS. d Anomalous Fourier map for the Fe-loaded PfFtn, contoured at 3.0 map RMS. e Anomalous Fourier map for the Fe-soaked PfFtn, contoured at 3.0 map RMS. f Anomalous Fourier map for the Zn-soaked PfFtn using the peak data, contoured at 3.0 map RMS. g Labeled view of the residues and metal sites in the Fe-soaked FC. h Anomalous Fourier map for the Zn-soaked PfFtn using the low-energy remote data, contoured at 3.0 map RMS. i Dispersive Fourier map for the Zn-soaked PfFtn, contoured at 2.8 map RMS. The anomalous electron-density maps were calculated using as amplitudes the anomalous difference coefficients obtained from each dataset, and the phases (rotated by 90°) obtained from the respective final structure refinement, except for the “Fe-loaded” data, for which the phases were taken from the “as-isolated” refinement. The Zn-dispersive Fourier map was calculated by first scaling together the peak and low-energy remote datasets with CCP4 SCALEIT [16] and then using as coefficients the difference F(low-energy remote) − F(peak) and the phases from the final “Zn-soak” refinement. a–c show that the occupation of site B in the as-isolated crystal is negligible; d shows a small residual occupancy of site B in the Fe-loaded structure and no evidence of site C occupancy; e shows a site B occupancy clearly lower than those of sites A and C in the Fe-soaked structure; f, h and i show that in the Zn-soaked structure, part of the originally present Fe in site A has been displaced to site C by the Zn ions, that site B is occupied by Zn only, and that site C is very likely occupied by Fe only. All panels were drawn with the same orientation. In all panels except g the bulk of PfFtn monomer is represented as a gray tube onto which the side chains (including Cα atoms) of the FC residues have been overlaid in ball-and-stick representation (carbon atoms blue-gray, oxygen atoms red, nitrogen atoms blue, iron atoms pink and zinc atoms cyan). In g, only the residues near the FC are represented in ball-and-stick mode, and are labeled for easier identification of the residues mentioned in the text. The figure was prepared with DINO [52]
For the Fe-loaded crystal structure, the anomalous difference Fourier map, calculated with the phases obtained from the final “as-isolated” refinement (Fig. 7d), shows a very small occupancy of site B. This map is very similar to that obtained from the MR phases (not shown). In the Fe-soaked crystal, the anomalous difference Fourier map calculated at the end of the refinement showed that sites A and C have a clearly higher Fe occupancy than site B (Fig. 7e).
In the Zn-soaked crystal, the anomalous difference Fourier maps obtained from the two datasets collected (just above and 176 eV below the Zn K-absorption edge) clearly show some iron occupation of sites A and C (Fig. 7h), whereas zinc populates sites A and B (Fig. 7f, i). The dispersive Fourier map calculated using the two datasets (Fig. 7i) shows Zn peaks at sites A and B but since no diffraction data were recorded at the inflexion point of the Zn K-absorption edge, this map is noisier and no conclusions can be drawn about any Zn occupation of site C. Since the as-isolated structure only showed Fe occupation of FC site A, we conclude that the Fe occupation of site C in the Zn-soaked structure results from iron initially present in site A, displaced to site C by the Zn soak. This is an interesting new result, since the previous Zn-soaked and Fe-soaked structures reported for AfFtn [2] and EcFtnA [1] were obtained from apoferritin crystals.
FC stability
Interestingly, while mutagenesis studies have shown both sites A and B to be important for iron uptake and oxidation in ferritins [38], in the crystallized proteins the occupancy of the both sites was found only in BFR from D. desulfuricans ATCC 27774 [31]. The PfFtn FC followed the common trend, and as mentioned already the occupation of FC sites B and C was only observed in Fe-soaked or Zn-soaked crystals. This contradicted our EPR studies that showed a fully developed dinuclear iron FC EPR signal in the sample prior to crystallization [39]. In order to check the integrity of the PfFtn FC, we loaded three samples of apo PfFtn with iron and incubated them for 1 day and 2 months at 4 °C in air and, mimicking the crystallization conditions, for 2 months in a 2 M ammonium sulfate solution at room temperature. The FC EPR signal from each sample was measured after titration to 130 mV, which is the potential at which the mixed-valence PfFtn EPR signal is maximum [39]. The amplitude of the FC signal from the sample incubated for 2 months in air was noticeably but not drastically smaller than that of the sample incubated for 1 day (Fig. 8). However, the FC EPR signal amplitude of the sample incubated for 2 months in a 2 M ammonium sulfate solution was fivefold smaller than that from the sample incubated for 2 months in air (Fig. 8). The coordination distance from Glu130 to site B (Table 5) is quite long (more than 2.8 Å); therefore, site B may be rather labile, which could account for its lower occupation in the Fe-soaked and Zn-soaked crystals, as well as for the easy removal of its iron atom by the precipitating agent ammonium sulfate during crystallization of the as-isolated ferritin.
Fig. 8.
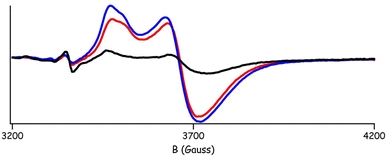
The Fe(III)–O–Fe(II) FC electron paramagnetic resonance (EPR) signal of 6 μM PfFtn 24-mer, titrated to 130 mV. The decrease in the amplitude of the signal depending on incubation time and the presence of crystallization solution in the sample illustrates the stability of the center. Prior to EPR measurement, the three samples tested were incubated, respectively, for 1 day after loading with iron (blue line), for 2 months (red line) and for 2 months in the presence of a crystallization solution, 2 M ammonium sulfate (black line)
There is an ongoing discussion in the literature on whether the FC iron sites A and B are transient in the process of iron uptake–oxidation–translocation into the protein core, or whether they form a stable cofactor [38]. In the current experiment, the decrease in the amplitude of the FC EPR signal in the sample incubated in air for 2 months appears to be too small to be caused by iron translocation into the protein cavity. Instead, as shown by the sample incubated for 2 months in ammonium sulfate, the most likely cause for the lack of the B iron site in the as-crystallized PfFtn is iron complexation and removal by the crystallization agent.
Thermostability
The thermal stability of a protein can often be described as an equilibrium between the protein’s folded and unfolded state, and frequently correlates with the growth temperature of the organism. Analysis of thermostability factors is often made via comparison of the genome/protein sequences from the hyperthermophiles, thermophiles and mesophiles, or via mutagenesis. In the literature, a number of factors have been proposed to be involved in thermostability. These include salt bridges, hydrogen bonds, van der Waals interactions, distribution of amino acids, higher β-strand content, more charged amino acids, fewer uncharged polar amino acids, more hydrophobic β-branched amino acids and others [40–46]. However, no single consistent set of determining factors has emerged from the different studies that have been carried out to date; therefore, it appears that protein thermal stability is the cumulative result of various factors.
Ferritins are large and highly symmetrical agglomerates of 24 single subunits. This suggests that these proteins possess forces that keep the big structure together. Horse spleen ferritin has been shown to resist inactivation by up to 10 M urea [47] and temperatures up to 93 ºC [48].
PfFtn has been shown to be an extremely thermostable protein. No melting temperature could be found with differential scanning calorimetry up to 120 °C and the protein could withstand incubation at 100 °C for 1 day or autoclaving at 120 °C for half an hour without loss of its iron-uptake activity [13]. There are no data in the literature regarding the thermostability of other thermophilic and hyperthermophilic ferritins.
In our analysis of PfFtn thermostability (summarized in Table 6), and given that no thermostability data are available in the literature on ferritins from microorganisms other than P. furiosus, we assumed that it is the most thermostable ferritin among those shown in Table 6, and we used the optimal growth temperature of the organism as a measure of relative thermostability for the other ferritins listed in Table 6. However, it should be noted that the denaturation temperature of a protein may be higher than the optimal growth temperature of the organism where it is produced.
Table 6.
Hydrogen bonds and salt bridges in the PfFtn 24-mer and its closest structural homologues, calculated with HBPLUS
| PDB ID | Source organism a | Total H-bonds b | MM | MS | SS | Salt bridges c | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intramolecular | Intermolecular | Intramolecular | Intermolecular | Intramolecular | Intermolecular | Intramolecular | Intermolecular | |||
| – | Pyrococcus furiosus (100 °C) | 4,630 | 3,377 | 24 | 439 | 72 | 544 | 174 | 172 | 67 |
| 1vlg | Thermotoga maritima (80 °C) | 4,767 | 3,237 | 30 | 540 | 117 | 633 | 210 | 189 | 120 |
| 1s3q | Archaeoglobus fulgidus (80 °C) | 4,457 | 2,788 | 24 | 412 | 80 | 453 | 176 | 114 | 134 |
| 1krq | Campylobacter jejuni (42 °C) | 4,632 | 3,360 | 24 | 648 | 72 | 384 | 144 | 72 | 72 |
| 1eum | Escherichia coli (37 °C) | 4,720 | 3,320 | 24 | 836 | 32 | 412 | 96 | 60 | 4 |
MM hydrogen bonds between main-chain atoms only, MS hydrogen bonds between main-chain and side-chain atoms, SS hydrogen bonds between side-chain atoms only
aOptimal growth temperature in parentheses
bCalculated using default program parameters
cA subset of SS hydrogen bonds, considering interactions between side chains of negatively charged Asp/Glu residues with side chains of positively charged Arg/His/Lys residues
The hydrogen bonds were calculated with HBPLUS [49] using default parameters, and were divided into three classes: between main-chain atoms only (MM); between main-chain and side-chain atoms (MS); and between side-chain atoms only (SS). The salt bridges were determined as a subset of SS hydrogen bonds, considering interactions between side chains of negatively charged Asp/Glu residues with side chains of positively charged Arg/His/Lys residues. Furthermore, a distinction between intramolecular (within the same monomer) and intermolecular (between neighboring monomers) hydrogen bonds was made. The MM bonds are mostly intramolecular and are responsible for the stability of the secondary structure elements (α-helices). The intramolecular MS and SS hydrogen bonds contribute to stabilize the 3D fold of the monomeric subunits. The intermolecular MS and SS hydrogen bonds are those that contribute most to the stability of the 24-mer.
A surprising result of this calculation was that the total number of hydrogen bonds in the ferritin 24-mers was quite similar. If we exclude the lowest number (4,457 for AfFtn) which can possibly be attributed to its unusual quaternary structure, PfFtn actually has the lowest total number of hydrogen bonds, two fewer than the ferritin from Campylobacter jejuni (CjFtn), 90 fewer than EcFtnA and 137 fewer than TmFtn. There is no clear single trend in the data listed in Table 6, which only emphasizes the complex nature of the structural factors governing thermostability, even within a group of closely related structures. However, a careful inspection shows some interesting features, which may have a bearing on the structural basis for the different thermostabilities of the different ferritins listed in Table 6. The most striking single feature in Table 6 is the marked increase in intramolecular salt bridges between the ferritins of mesophilic (EcFtnA and CjFtn) and hyperthermophilic (AfFtn, TmFtn and PfFtn) organisms. Indeed, a similar but less clear trend is observed for the SS hydrogen bonds overall. These structural features could explain the higher resistance to thermal inactivation in the latter. However, this trend by itself does not validate our initial assumption that PfFtn is the most thermostable ferritin in the group. A secondary trend can be perceived for the main-chain intramolecular hydrogen bonds in the hyperthermophilic organisms (AfFtn, TmFtn and PfFtn), which is greatest in PfFtn, and may offset the effect of a lower number of intramolecular salt bridges in comparison with TmFtn. Therefore, the most important factor that protects PfFtn and other ferritins from thermal denaturation appears to be the preservation of the monomer fold, rather than the 24-mer assembly.
Conclusions
In this study, we reported the crystal structure of PfFtn. High sequence similarity between PfFtn, TmFtn and AfFtn suggests a separate group of thermostable ferritins. PfFtn is the second archaeal ferritin structure to be reported, following that of AfFtn. In spite of the high sequence identity (50%) between these two proteins, their quaternary structures are significantly different. PfFtn exhibited the 432 symmetry commonly found in other known ferritins, whereas AfFtn showed the 23 symmetry similar to dodecameric DPS proteins. The 23 symmetry of the AfFtn 24-mer resulted in four triangular pores with an approximate size of 45 Å, making large openings in the AfFtn shell. Therefore, the 23 symmetry of the AfFtn 24-mer remains an exception among ferritins, and further research is required to explain such an unusual conformation.
Three iron sites were observed in the FC of PfFtn monomer. Site A was present in the as-crystallized protein and sites B and C were observed only upon crystal soaking with either Fe or Zn ions. Combining these results with our measurements of the FC EPR signal prior to crystallization, where samples were incubated in air for 1 day and 2 months, and for 2 months in 2 M ammonium sulfate (the crystallization solution), we conclude that the lack of iron atoms at the B site is very likely due to their complexation by ammonium sulfate, and that the absence of these sites in the as-crystallized protein cannot be taken per se as evidence of a transient nature of the FC, in PfFtn and other ferritins.
The possible iron entry and exit routes of PfFtn, the threefold and fourfold channels, are similar to those from bacterial ferritins in the sense that the threefold channel is less hydrophilic and the fourfold channel is more polar than the channels of mammalian H and L chain ferritins.
The hyperthermostability of PfFtn was analyzed at the structural level by comparing the number of salt bridges and hydrogen bonds in PfFtn with those in AfFtn, TmFtn, CjFtn and EcFtnA. Although there are no data on the thermal stability of other thermostable ferritins, our results suggest that TmFtn, the ferritin from T. maritima, a bacterium which grows optimally at 80 °C, is probably more thermostable than PfFtn. Owing to a high number of intramolecular hydrogen bonds between main-chain atoms, and between main-chain and side-chain atoms, the main factor contributing to the high thermostability of PfFtn as well as the other structurally similar ferritins from hyperthermophilic microorganisms appears to be the preservation of the monomer fold rather than the 24-mer structure.
Acknowledgments
The authors are grateful to Ian Tickle (Astex Therapeutics, Cambridge, UK) for helpful suggestions and to Emile Bol (Delft University of Technology, Delft, The Netherlands) for advice on PyMOL. The authors would like to acknowledge the staff of the EMBL Grenoble Outstation, in particular Martin Walsh and Hassan Belrhali for technical assistance with the measurements at the EMBL-CRG BM 14 beamline. Financial support is acknowledged from the European Union—Research Infrastructure activity under the FP6 “Structuring the European Research Area” I3 program and from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (CW-NWO) under project no. 700.51.301.
Contributor Information
Jana Tatur, Email: J.Tatur@tnw.tudelft.nl.
Wilfred R. Hagen, Email: w.r.hagen@tnw.tudelft.nl
Pedro M. Matias, Phone: +351-21-4469669, FAX: +351-21-4463644, Email: matias@itqb.unl.pt
References
- 1.Stillman TJ, Hempstead PD, Artymiuk PJ, Andrews SC, Hudson AJ, Treffry A, Guest JR, Harrison PM (2001) J Mol Biol 307:587–603 [DOI] [PubMed]
- 2.Johnson E, Cascio D, Sawaya MR, Gingery M, Schröder I (2005) Structure 13:637–648 [DOI] [PubMed]
- 3.Stiefel EI, Watt GD (1979) Nature 279:81–83 [DOI] [PubMed]
- 4.Crichton RR (2001) Inorganic biochemistry of iron metabolism: from molecular mechanisms to clinical consequences. Wiley, Chichester
- 5.Webb J, Macey DJ, Talbot V (1985) Arch Environ Contam Toxicol 14:403–407 [DOI]
- 6.Lonnerdal B, Bryant A, Liu X, Theil EC (2006) Am J Clin Nutr 83:103–107 [DOI] [PubMed]
- 7.Jacobs A, Worwood M (eds) (1980) Iron in biochemistry and medicine, vol 2. Academic, London
- 8.Powers JM (2006) J Neuropathol Exp Neurol 65:716–721 [DOI] [PubMed]
- 9.Takahashi T, Kuyucak S (2003) Biophys J 84:2256–2263 [DOI] [PMC free article] [PubMed]
- 10.Zeth K, Offermann S, Essen LO, Oesterhelt D (2004) Proc Natl Acad Sci USA 101:13780–13785 [DOI] [PMC free article] [PubMed]
- 11.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM (1998) Nat Struct Biol 5:294–303 [DOI] [PubMed]
- 12.Ilari A, Stefanini S, Chiancone E, Tsernoglou D (2000) Nat Struct Biol 7:38–43 [DOI] [PubMed]
- 13.Tatur J, Hagedoorn PL, Overeijnder ML, Hagen WR (2006) Extremophiles 10:139–148 [DOI] [PubMed]
- 14.Matias PM, Tatur J, Carrondo MA, Hagen WR (2005) Acta Crystallogr Sect F 61:503–506 [DOI] [PMC free article] [PubMed]
- 15.Leslie AGW (1992) Joint CCP4 ESF-EACMB Newsl Protein Crystallogr 26:27–33
- 16.Collaborative Computational Project Number 4 (1994) Acta Crystallogr Sect D 50:760–763 [DOI] [PubMed]
- 17.Storoni LC, McCoy AJ, Read RJ (2004) Acta Crystallogr Sect D 60:432–438 [DOI] [PubMed]
- 18.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS (2001) Acta Crystallogr Sect D 57:1445–1450 [DOI] [PubMed]
- 19.Cowtan K (1994) Joint CCP4 ESF-EACBM Newsl Protein Crystallogr 31:34–38
- 20.Roussel A, Fontecilla-Camps JC, Cambillau C (1990) XV IUCr Congress Abstracts, Bordeaux, France, pp 66–67
- 21.Murshudov GN, Vagin AA, Dodson EJ (1997) Acta Crystallogr Sect D 53:240–255 [DOI] [PubMed]
- 22.Schomaker V, Trueblood KN (1968) Acta Crystallogr Sect B 24:63–76 [DOI]
- 23.Lamzin VS, Wilson KS (1993) Acta Crystallogr Sect D 49:129–147 [DOI] [PubMed]
- 24.Brünger AT (1992) Nature 355:472–474 [DOI] [PubMed]
- 25.Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) J Appl Crystallogr 26:283–291 [DOI]
- 26.Ramachandran GN, Sasisekharan V (1968) Adv Protein Chem 23:283–438 [DOI] [PubMed]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242 [DOI] [PMC free article] [PubMed]
- 28.Emsley P, Cowtan K (2004) Acta Crystallogr Sect D 60:2126–2132 [DOI] [PubMed]
- 29.Tickle IJ, Laskowski RA, Moss DS (1998) Acta Crystallogr Sect D 54:547–557 [DOI] [PubMed]
- 30.Kabsch W, Sander C (1983) Biopolymers 22:2577–2637 [DOI] [PubMed]
- 31.Macedo S, Romao CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA (2003) Nat Struct Biol 10:285–290 [DOI] [PubMed]
- 32.Yu B, Blaber M, Gronenborn AM, Clore GM, Caspar DL (1999) Proc Natl Acad Sci USA 96:103–108 [DOI] [PMC free article] [PubMed]
- 33.Krissinel E, Henrick K (2004) Acta Crystallogr Sect D 60:2256–2268 [DOI] [PubMed]
- 34.Levi S, Santambrogio P, Corsi B, Cozzi A, Arosio P (1996) Biochem J 317:467–473 [DOI] [PMC free article] [PubMed]
- 35.Rice DW, Ford GC, White JL, Smith JMA, Harrison PM (1983) In: Theil EC, Eichhorn GL, Marzilli LG (eds) Advances in inorganic biochemistry, vol 5. Elsevier, New York, pp 39–50
- 36.Wardeska JG, Viglione B, Chasteen ND (1986) J Biol Chem 261:6677–6683 [PubMed]
- 37.Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, Cesareni G, Thomas CD, Shaw WV, Harrison PM (1991) Nature 349:541–544 [DOI] [PubMed]
- 38.Lewin A, Moore GR, Le Brun NE (2005) Dalton Trans 22:3597–3610 [DOI] [PubMed]
- 39.Tatur J, Hagen WR (2005) FEBS Lett 579:4729–4732 [DOI] [PubMed]
- 40.Vogt G, Woell S, Argos P (1997) J Mol Biol 269:631–643 [DOI] [PubMed]
- 41.Kannan N, Vishveshwara S (2000) Protein Eng 13:753–761 [DOI] [PubMed]
- 42.Eidsness MK, Richie KA, Burden AE, Kurtz DM Jr, Scott RA (1997) Biochemistry 36:10406–10413 [DOI] [PubMed]
- 43.Vetriani C, Maeder DL, Tolliday N, Yip KS, Stillman TJ, Britton KL, Rice DW, Klump HH, Robb FT (1998) Proc Natl Acad Sci USA 95:12300–12305 [DOI] [PMC free article] [PubMed]
- 44.Kumar S, Tsai CJ, Nussinov R (2000) Protein Eng 13:179–191 [DOI] [PubMed]
- 45.Chakravarty S, Varadarajan R (2000) FEBS Lett 470:65–69 [DOI] [PubMed]
- 46.Suhre K, Claverie JM (2003) J Biol Chem 278:17198–17202 [DOI] [PubMed]
- 47.Liu X, Jin W, Theil EC (2003) Proc Natl Acad Sci USA 100:3653–3658 [DOI] [PMC free article] [PubMed]
- 48.Stefanini S, Cavallo S, Wang CQ, Tataseo P, Vecchini P, Giartosio A, Chiancone E (1996) Arch Biochem Biophys 325:58–64 [DOI] [PubMed]
- 49.McDonald IK, Thornton JM (1994) J Mol Biol 238:777–793 [DOI] [PubMed]
- 50.DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos. http://www.pymol.sourceforge.net/
- 51.Combet C, Blanchet C, Geourjon C, Deleage G (2000) Trends Biochem Sci 25:147–150 [DOI] [PubMed]
- 52.Philippsen A (2003) http://www.dino3d.org


