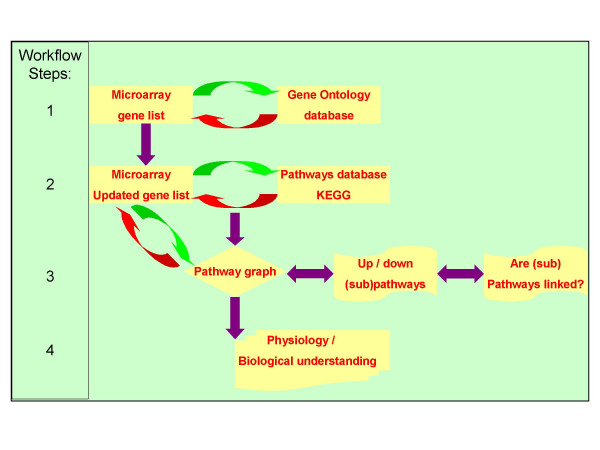
Figure 1.
The workflow diagram describing the individual steps taken by the software from microarray data to physiological understanding via pathways analysis. Step 1: A PERL script uses a text file with a list of all genes on the microarray to search the Gene Ontology database for synonyms. These Synonyms are added to the gene list. Step 2 uses this updated gene list to search the KEGG pathway database for pathways in which the genes are involved. If one or more pathways were found for a gene the KEGG database returns a list of pathway names for that gene and a link to the reference pathway for each pathway. Both are added to the file. Step 3 combines the results of the microarray and the pathways. All genes of the pathway represented on the microarray have an expression pattern consisting of the expression in the Longissimus muscle at seven time points during gestation. First all genes of the pathway are considered. Secondly, if more than one biochemical path is specified by the pathway (i.e. called subpathways) the individual subpathways are investigated separately. Thirdly, if KEGG-pathways are linked either because the pathway indicates it or because at least one gene is found in two or more pathways, a network of these pathways is constructed. In step 4 the expression patterns of these pathways and networks were analysed for comparable expression patterns that may indicate common regulatory events linking genes in pathways, subpathways, or networks of pathways creating biological understanding of the physiology of the studied processes.