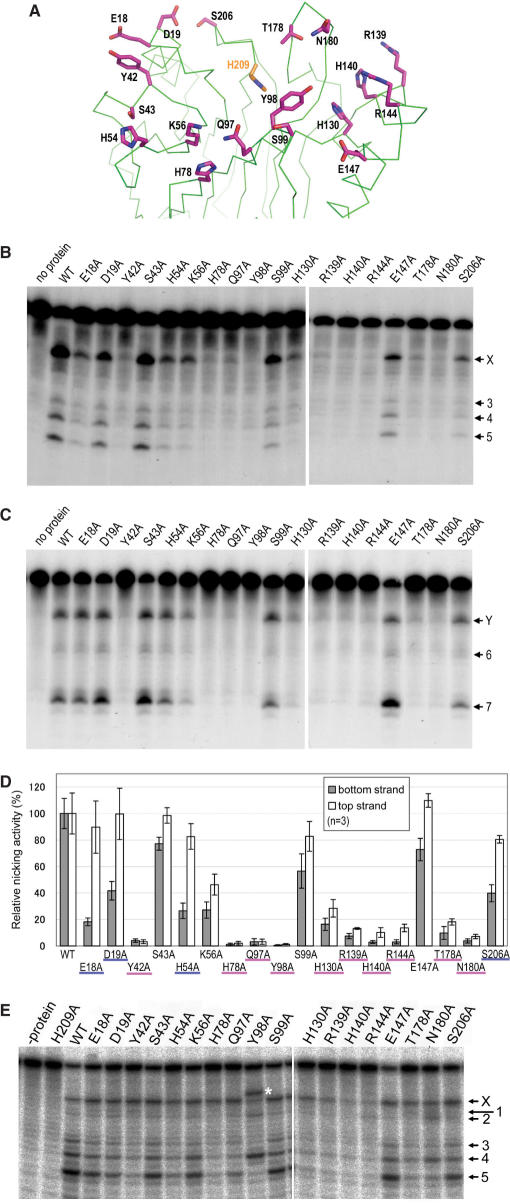
Figure 6.
Mutagenesis study of R1Bm EN. (A) Positions of residues mutated in this study. The positions of residues substituted with alanine are indicated as line models (magenta). His-209 is also depicted to indicate the catalytic site. (B) Bottom strand cleavage by R1Bm EN mutants. Three picomoles of the substrate was cleaved with wild-type R1Bm EN and the mutants. The solid arrowheads indicate the target site cleavage of the bottom strand. (C) Top strand cleavage by R1Bm EN mutants. One picomole of the substrate was used. The open arrowheads indicate the target site cleavage of the top strand. (D) Nicking activities of R1Bm EN and the mutants. The cleavage products nicked at the target site were quantified and the percentage of the cleavage product relative to that of wild-type R1Bm EN was shown in each strand cleavage. The mutants representing <20% of wild-type activity in both strand cleavage are underlined in magenta and the mutants showing the great reduction of the nicking activity only in bottom strand cleavage are underlined in blue. The results of three independent experiments were averaged and error bars show S.E. (E) Cleavage of the bottom strand by R1Bm EN mutants under the vigorous condition. The cleaved bands corresponding to Figure 1B are indicated. Asterisk indicates the abnormal band observed in Y98A mutant.