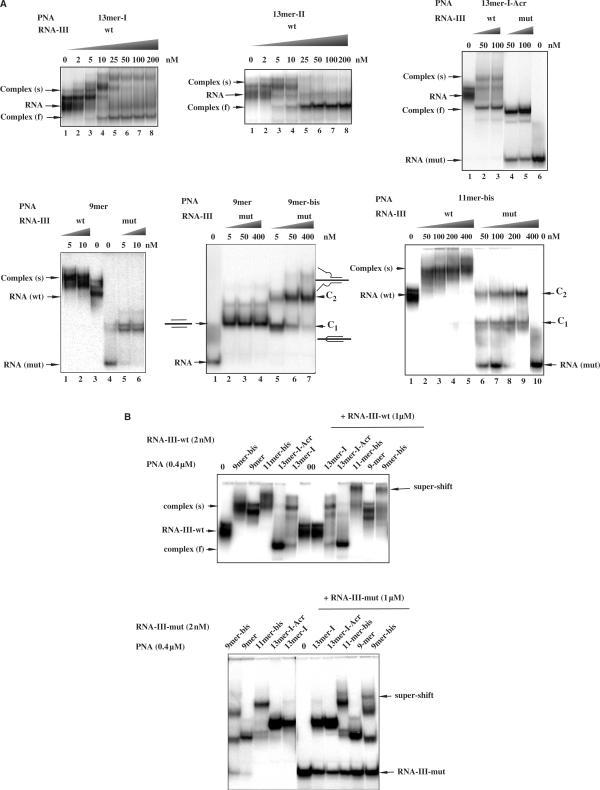
Figure 2.
(A) Gel mobility-shift analysis of PNAs binding to their target RNA. A fixed concentration of the 32P-labeled RNA-III-wt and mismatches containing RNA-III-mut (2 × 10−9 M) were incubated in 50 mM Tris (pH 8) buffer with increasing concentration of PNA as indicated above the lanes. Slow migrating complexes (s) correspond to the complexes formed with folded RNA whereas higher electrophoretic mobility complexes (f) correspond to the invasion complexes that unfold the structured RNA target. C1 and C2 denote complexes involving one and two molecules of bis-PNAs respectively. (B) An excess of the RNA target sequence (2.5-fold excess relative to PNA concentration (400 nM)) was added to the complexes and incubated for 5 min. Bound and unbound complexes were separated in a 15% non-denaturing polyacrylamide gel at 20°C. RNA-III-wt (top) and RNA-III-mut (bottom) were used as targets.