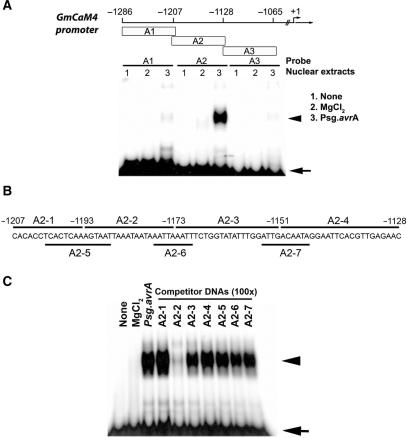
Figure 1.
Identification of a pathogen-responsive protein-binding site in the GmCaM-4 promoter. (A) Schematic diagram of DNA fragments derived from the GmCaM-4 promoter used for gel mobility shift assay (EMSA) and their patterns of binding with nuclear extracts prepared from W82 soybean suspension culture cells treated with MgCl2 (lanes 2, MgCl2) or Pseudomonas syringae pv glycinea carrying avrA (Psg. avrA) (lanes 3, Psg. avrA). None (lanes 1) represents free DNA probe. The positions of fragments upstream of the GmCaM4 transcriptional start site are indicated. (B) Oligonucleotides (A2-1–A2-7) corresponding to the GmCaM-4 promoter A2 region that were used as competitors. (C) Competitive gel mobility shift assay. The DNA binding reaction was performed by pre-incubating unlabeled competitors (A2-1–A2-7) and then adding the 32P-labeled A-2 fragment as a probe. A 100-fold molar excess of competitor DNA was added to each reaction mixture. Free (arrow) and protein-complexed (arrowhead) probes were separated on an 8% polyacrylamide gel and visualized by autoradiography.