Abstract
RNA-editing enzymes of the ADAR family convert adenosines to inosines in double-stranded RNA substrates. Frequently, editing sites are defined by base-pairing of the editing site with a complementary intronic region. The glutamate receptor subunit B (GluR-B) pre-mRNA harbors two such exonic editing sites termed Q/R and R/G. Data from ADAR knockout mice and in vitro editing assays suggest an intimate connection between editing and splicing of GluR-B pre-mRNA.
By comparing the events at the Q/R and R/G sites, we can show that editing can both stimulate and repress splicing efficiency. The edited nucleotide, but not ADAR binding itself, is sufficient to exert this effect. The presence of an edited nucleotide at the R/G site reduces splicing efficiency of the adjacent intron facilitating alternative splicing events occurring downstream of the R/G site.
Lack of editing inhibits splicing at the Q/R site. Editing of both the Q/R nucleotide and an intronic editing hotspot are required to allow efficient splicing. Inefficient intron removal may ensure that only properly edited mRNAs become spliced and exported to the cytoplasm.
INTRODUCTION
Genome sequencing projects of higher eukaryotes have revealed a surprisingly low number of genes that fail to explain their organismic and developmental complexity (1). Post-transcriptional processes that recode, diversify and fine tune the transcriptome are now regarded as the potential players leading to evolutionary variation. Alternative splicing and RNA editing are the key events leading to transcriptome diversification (2,3). Small non-coding RNAs, in turn, fine-tune RNA stability and translatability (4).
RNA editing by adenosine deaminases that act on RNA (ADARs) is widespread in metazoa (5). ADARs deaminate adenosines to inosines (A-to-I) within double-stranded or structured RNAs. As inosines resemble guanosines, editing by ADARs can alter splice sites or change the coding potential of an RNA and, therefore, generate diversity in proteins that are encoded by a single gene (3). Moreover, ADAR-mediated editing can affect non-coding sequences such as introns, UTRs or miRNAs thereby altering the secondary structure, stability or base-pairing potential of edited RNAs (6–9). Mammals have two active editing enzymes, ADAR1 and ADAR2, which exhibit different substrate specificities (10–13). A third protein, ADAR3, seemingly lacks enzymatic activity (14). ADARs contain a conserved deaminase domain and two or three double-stranded RNA binding domains (dsRBDs).
Both viral and cellular ADAR targets have been described. Most cellular substrates are found in the central nervous system, but also non-neuronal substrates are increasingly being discovered (15). A well-studied substrate for ADAR editing is the pre-mRNA encoding glutamate receptor subunit B (GluR-B) (16). This RNA is edited at two exonic sites termed the Q/R and R/G, respectively. Editing at these sites leads to codon exchanges and thus alters the properties of GluR-B-containing ion channels. Editing at the Q/R site, located in exon 11, changes a glutamine codon to an arginine codon and results in a lower Ca2+ permeability of the channel (17,18). Furthermore, editing at the Q/R site is essential for proper tetramer assembly of AMPA receptors (19). The R/G site is located in exon 13 upstream of an alternatively spliced region known as the flip/flop module (20). Here, an arginine codon is converted to a glycine codon, which allows faster recovery of the receptor from desensitization (21). Two additional editing sites are found in intron 11, called hotspot 1 (or +60 site) and hotspot 2 (or+262/263/264 site), respectively (22). The Q/R site and hotspot 2 are solely edited by ADAR2, whereas hotspot 1 and the R/G site can be edited by ADAR1 and ADAR2 (23,24).
Editing at the Q/R site is nearly complete. Mice lacking ADAR2 are prone to epileptic seizures and die about three weeks after birth; however, they can be completely rescued by introducing a ‘pre-edited’ GluR-B gene (23). Editing levels at the R/G site reach 75% in adult mice, but are much lower in embryos and gradually increase throughout development (21).
In most coding targets, exonic editing sites depend on an intronic editing complementary sequence (ECS) (22). The ECS, base-pairs with the editing site to form the double-stranded structure required for ADAR binding. Editing must, therefore, be a co-transcriptional event that occurs prior to intron removal (25).
The close proximity of editing and splice sites coupled with experimental data suggest that RNA editing and splicing are coordinated in vivo: for instance, mice lacking ADAR 2 are deficient in the removal of intron 11 in GluR-B pre-mRNA which neighbors the Q/R editing site (23). Coordination of editing and splicing has also been observed at the R/G site of GluR-B pre-mRNA. Here ADAR2 inhibits splicing in vitro, but seemingly not in vivo (26). The C-terminal domain of RNA Pol-II might play a role in coordinating editing at nascent transcripts as it is required for efficient autoediting of ADAR2 pre-mRNA, but not for splicing (27). Moreover, in malignant gliomas, hypoediting correlates with alternative splicing in 5-HT2C serotonin receptors (28). Finally, in Drosophila, a strong correlation between alternative splice site choice and editing efficiency upstream of the alternative splice site was observed for two ADAR substrates (29).
In principle, RNA editing could affect splicing via three alternative mechanisms: first, editing might alter cis-acting signals that modulate splicing activity. Second, editing could destabilize a double-stranded structure, thereby allowing access for proteins to the splice site. Third, ADAR binding, but not editing itself, could help to recruit factors that regulate splicing.
To distinguish amongst these possibilities, we investigated the correlation between splicing and editing at the R/G and Q/R sites in GluR-B pre-mRNA using a novel cell-based splice assay. Reporter constructs containing the editing sites and constructs that either mimic edited RNA, or in which editing and/or ADAR binding is prevented, were tested. Our results show that editing at the R/G site leads to a reduction in splicing efficiency in the adjacent intron and influences the downstream alternative splicing event. This phenomenon is caused by the inosine at the R/G site and does not require ADAR binding. Furthermore, editing of both the Q/R nucleotide and an intronic editing hotspot is required for efficient splicing of the intron adjacent to the Q/R site.
MATERIALS AND METHODS
Construction of RFP and GFP expression plasmids
The EGFP ORF was amplified from pEGFP-C2, (Clontech) by PCR using suitable primers and cloned in-frame, downstream of the RFP ORF in dsRed express. To separate the two tags, a flexible linker (an unstructured region of the β-galactosidase gene) was inserted between the two ORFs. Additionally, a nuclear localization signal (SV40 large T-antigen NLS) and a polylinker was inserted into the expression plasmid. This vector was the starting construct into which further DNA fragments for investigation were introduced. As a splice control, the adenovirus major late pre-mRNA (Ad1), was introduced (30). An unspliceable Ad1 pre-mRNA was produced by mutating the 5′ and 3′ splice consensus sequences of the Ad1 intron using the method described in the QuickChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA,USA).
Generation of GluR-B constructs
GluR-B sequences were amplified by PCR from mouse genomic DNA. For sequences spanning exons 13 through 14, the following primers were used: 5′ TCGAGAATTCTTGCAGTGTTTGATAAAATGTGGA 3′ (forward, containing an EcoRI site) and 5′ CTTCGGTACCCACTCTCCTTTGTCGTACCACCA 3′ (reverse, containing a KpnI site). A ‘pre-edited’ version was generated by site-directed mutagenesis. An ‘uneditable’ version was produced by replacing the sequence of the ECS, thus preventing formation of a double-stranded RNA. To create an uneditable ‘binding but not editing’ R/G-containing construct, the cytosine in the ECS opposing the edited base was mutated to a guanosine (31). A GluR-B DNA stretch that spans exons 13 through 16 was amplified using primers: 5′ TGTAGTCGACAATTGCAGTGTTTGATAAAA 3′ (forward) and 5′ ATAGGTACCTTAACACTCTCGATGCCATA 3′ (reverse). The Q/R editing constructs spanning exons 11 through 12, were amplified with primers: 5′ TCTTGTCGACGAGCCTTGGAATCTCTATCATG 3′ (forward) and 5′ AGGAGGATCCAACTCTTTAGTGGAGCCAGAGT 3′ (reverse). ‘Pre-edited’ variants of the Q/R site and intronic hotspot 2, respectively, were introduced by site-directed mutagenesis. Deletion mutants were created using internal restriction sites. All inserts were controlled by sequencing.
Cell culture and transfection
HeLa and HEK293 cell lines were transfected using either Lipofectamine 2000 (Invitrogen, CA,USA) or Nanofectin (PAA, Pasching, Austria) according to the manufacturer's instruction. Cells were analyzed 24–48 h post transfection.
RNA isolation and RT-PCR
RNA was isolated with Trifast reagent (PEQLAB, Erlangen, Germany) followed by two rounds of DNAseI digestion. Additionally, to remove residual transfected DNA, the sample was digested with DpnI. RevertAid M-MuLV Reverse Transcriptase (Fermentas, Lithuania) was used for reverse transcription using a GFP-specific primer: 5′ CCTCTACAAATGTGGTATGGCTG 3′. The 1/10th of the reactions was used for PCR reactions. Plasmids that were used for the transfections were used as PCR controls. The following primers were used for the amplifications: 5′ GGTGGAGTTCAAGTCCATCTACATGG 3′ (forward, in RFP); 5′ TCGACCAGGATGGGCACCAC 3′ (reverse, in GFP); 5′ ACCTCATATCCGTATACAAACCGTT 3′ (reverse, in intron 13 of mouse GluR-B); 5′ GCAGCAAGCTTGACAACAAAAA 3′ (reverse, in Ad1 intron); 5′ GCAGCTGCTGACATCTTTATAGTG 3′ (reverse, in intron 11 of mouse GluR-B).
Amplification and sequencing of GluR-B cDNAs from mouse brain
The following primers were used for amplification: 5′ TCTTGTCGACGAGCCTTGGAATCTCTATCATG 3′ (forward, exon 11); 5′ ATATGGATCCGTGGCGATGCCGTAGCCTTTGGAA 3′ (reverse, exon 12); 5′ TGTAGTCGACAATTGCAGTGTTTGATAAAA 3′ (forward, exon 13 with a SalI site) and 5′ ATAGGTACCTTAACACTCTCGATGCCATA 3′ (reverse, exon 16 with a KpnI site). PCR products were purified, cloned into the pCR2.1 vector (Invitrogen, CA,USA) and individual clones were sequenced.
Microscopic analysis
Cells grown on coverslips were fixed and stained as previously described (32). Measurements of fluorescence intensities were carried out with the help of Quantity One software (Biorad, CA,USA).
FACS analysis and statistical analysis
The flow cytometry data was collected on a FACScalibur (Becton and Dickinson, NJ, USA) using the CellQuest 3.3 software. The data was further analyzed with the FlowJo 6.3.1 software. To calculate splicing efficiencies, cells with moderate red fluorescence were gated and the fluorescence intensities in the red (constitutively expressed) and green (only visible after splicing) channels were collected. To determine splicing efficiencies, the ratio of green (spliced) to red (constitutive) fluorescence was calculated for each gated event. At least 1800 events were collected for each sample. The mean and SD was calculated for each sample set. For clarity, the mean of the ratio of green to red fluorescence was set to 1 in the RNLG sample which expresses both RFP and GFP constitutively. All other values were normalized accordingly.
To determine whether variations amongst different samples are statistically significant. A student's t-test was performed for all data sets using Microsoft Excel.
RESULTS
The reporter assay
To investigate the effect of RNA editing on splicing of the GluR-B pre-mRNA, a splice assay was established that allows the quantification of splicing efficiencies in vivo. cDNAs encoding red and green fluorescent proteins (RFP and GFP) were cloned in-frame into a tissue culture expression vector separated by a flexible region of the Escherichia coli lac Z gene, an NLS and a polylinker. Genomic fragments containing splicing and editing sites and their flanking introns were then inserted into the polylinker separating the RFP and GFP open reading frames (ORFs) (Supplementary Figure S1, Figure 1). Upon transfection into tissue culture cells the RFP reporter was constitutively expressed, while expression of the GFP reporter depended on the removal of the intron. As a positive control, constructs were used that contained either only the flexible linker region between the RFP and GFP reporter, or a standard splicing substrate, the adenovirus major late pre-mRNA (Ad1). To further control for the effect of nonsense mediated decay (NMD) on RFP expression, a splicing-deficient Ad1 was also tested.
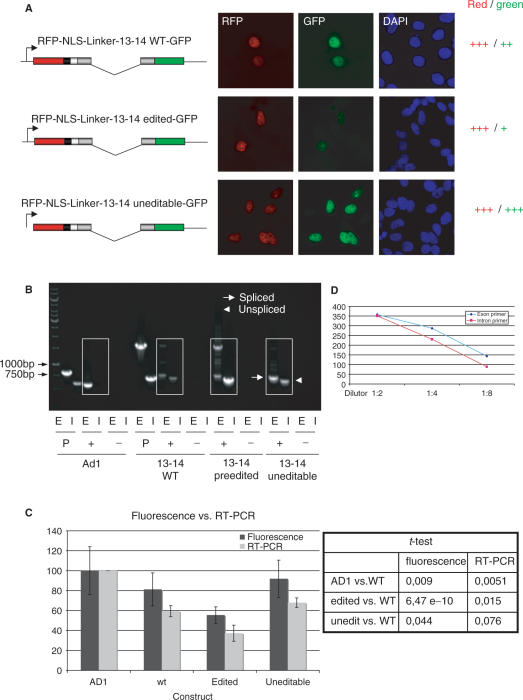
Figure 1.
Analysis of splicing efficiencies of wild-type and mutant R/G constructs. (A). Microscopic images of HeLa cells transfected with wild-type, ‘pre-edited’, and uneditable R/G constructs. The constructs are indicated next to each picture series. RFP and GFP fluorescence is shown in red and green, respectively. DNA is stained with DAPI (blue). A ‘pre-edited’ R/G shows reduced fluorescence in the GFP channel when compared to wild-type or uneditable R/G constructs, suggesting impaired splicing efficiency. Scale bar: 10 µm. (B) RT-PCR analysis of cells transfected with Ad1, wild-type R/G, ‘pre-edited’ R/G, and uneditable R/G fragments. Exonic (E) and intronic (I) primers were used to selectively amplify spliced and unspliced products. Arrow: PCR product derived from the spliced product; Arrowhead: PCR product derived from the unspliced product. Constructs are indicated below the respective lanes. P: plasmid positive control; +: with RTase; −: without RTase. To allow quantification of PCR products and to avoid saturation of the PCR reactions, only 23 cycles were run. The high molecular band obtained with exonic primers in the pre-edited construct reflects the unspliced primary RNA and results from the poor splicing of this construct. (C) Bar diagram representing the averaged results of 3 RT-PCR experiments (light gray bars) and 100 fluorescent cells analyzed by quantitative microscopy (dark bars). In both assays, a ‘pre-edited’ R/G-containing fragment (edited) showed reduced splicing. Splicing efficiency of Ad1 was set to 100%. A student's t-test indicates that the observed differences are significant (P-value < 0.05). (D) Quantified band intensities of RT-PCRs with different DNA dilutions are plotted. The linear relationship between the different DNA dilutions are shown for each primer pair. A 1:4 dilution was used for the RT-PCR in (B).
Microscopical analysis of transfected HeLa cells (Supplementary Figure S1) revealed that the vector containing the two reporters separated by the flexible linker, gave strong, evenly distributed nuclear signals in the red and green channels. The splice control, Ad1, showed the same result, indicating splicing of the Ad1 pre-mRNA. The splicing-deficient Ad1, on the other hand, only displayed red fluorescence, showing that neither NMD nor ‘leak-through’ of signals from the red into the green channel interfere with the readout of the assay.
A ‘pre-edited’ R/G construct shows reduced splicing
Fragments of mouse GluR-B spanning the majority of exon 13, containing the R/G site, the adjacent intron 13 and a large piece of exon 14 were introduced into the reporter vector (mimicking ‘wild type’) and tested for splicing efficiency. A ‘pre-edited’ construct was generated by inserting a guanosine at the site of editing. To generate an uneditable construct, the sequence of the ECS was changed, preventing formation of a target editing site (Supplementary Figure S2). In HeLa cells, which show moderate levels of editing, the uneditable mutant exhibited the strongest GFP fluorescence when compared to RFP fluorescence (Figure 1A). The wild-type construct displayed intermediate green signals while the ‘pre-edited’ R/G construct showed the weakest green signals. This result suggests that splicing of the mini-reporter construct is inhibited by editing. The wild-type construct, which can be edited by endogenous ADAR, showed a slight reduction in splicing while the ‘pre-edited’ construct was spliced least efficiently.
For quantification, 100 cells were recorded in the green and red channels, respectively, and fluorescence ratios were calculated (Figure 1C). Furthermore, splicing efficiencies were quantified by RT-PCR using sets of primers specific for intronic (unspliced) and exonic (spliced) regions (Figure 1B and 1D). In both assays (fluorescence of 100 cells versus RT-PCR experiments), the ‘pre-edited’ construct showed reduced splicing efficiency, while an uneditable construct was most efficiently spliced. Student's t-test indicated that the observed differences between wild-type and pre-edited or the uneditable construct were significant. Most interestingly both methods give similar results, indicating that either technique can be used to measure splicing efficiencies.
Inosine at the R/G site is sufficient to reduce splicing efficiency
To allow faster and more quantitative measurements of splicing efficiencies, we employed fluorescence activated cell sorting (FACS) to determine fluorescence intensities (Figure 2).
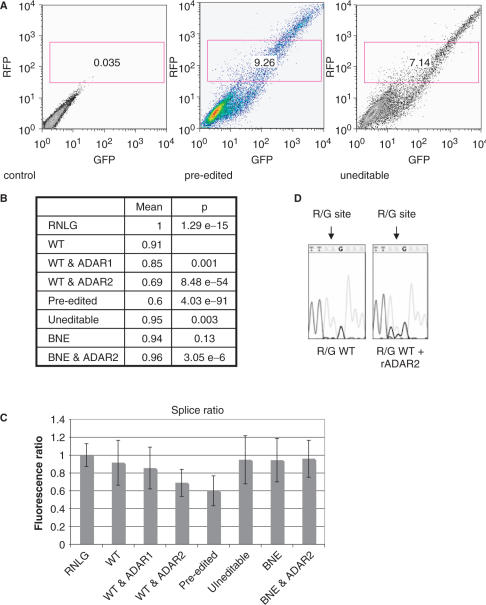
Figure 2.
FACS analysis of R/G constructs transfected into HEK293 cells. (A) 2D plot of untransfected cells (control), cells transfected with a vector containing exons 13 through 14 including the R/G site in a pre edited, and uneditable state. Constitutive red fluorescence is plotted along the y-axis while green fluorescence (only visible after successful splicing) is plotted along the x-axis. Only cells expressing solid RFP expression were chosen and gated for further analysis. (B) Red to green fluorescence ratios of individual cells in the gated window shown in (A) were calculated. The mean of these ratios is given. For clarity, the mean fluorescence ratio of cells transfected with ‘empty vector’ (RNLG) was set to 1. Wild-type and mutant R/G constructs spanning exons 13 through 14, and plasmids expressing ADAR1 or ADAR2 are shown. A ‘pre-edited’ R/G construct and a wild-type construct cotransfected with ADAR2 are weakly spliced. However, a construct where ADAR can bind but is unable to edit (binding but no editing +ADAR2) showed GFP fluorescence comparable to the wild-type construct, indicating that an inosine at the R/G site and not ADAR binding is responsible for splice suppression. Student's t-test P-values indicate that populations of transfected cells differed significantly from cells transfected with the wild-type construct with the exception of cells transfected with the construct that could be bound but not edited by ADAR2 (BNE). (C) Bar diagram and SD of RFP:GFP fluorescence ratios shown in (B). (D) Cotransfection of ADAR2 results in editing of GluR-B at the R/G site. Sequencing of RT-PCR products revealed that wild-type R/G fragments display no G peak at the R/G site whereas cotransfection of ADAR 2 results in editing. The R/G site is marked by an arrow.
HEK293 cells, which have low endogenous editing activity, were used for subsequent cotransfection experiments (33). Wild-type, ‘pre-edited’, and uneditable R/G constructs were cotransfected with plasmids expressing either ADAR1 or ADAR2, or an empty control plasmid. After 48 h red and green fluorescence of approximately 20 000 cells were analyzed by FACS. The gate was set to analyze cells that exhibited moderate red fluorescence (Figure 2A: red fluorescence, y-axis; green fluorescence, x-axis). Within the gate, the ratio of green (spliced) to red (input) fluorescence was calculated for each event. The mean and SD of these ratios is given in 2B and is graphically displayed in 2C. These experiments confirm that a ‘pre-edited’ R/G fragment is spliced less efficiently than a wild-type or uneditable construct (Figure 2B and C). Cotransfection of a wild-type construct with ADAR2 reduces splicing efficiency, suggesting that ADAR2 edits the R/G site. Bulk sequencing of RT-PCR products of transfected cells confirmed editing at the R/G site in the presence of ADAR2 (Figure 2D).
To test whether binding of ADAR2 to the R/G site had an effect on splicing, we tested a GluR-B mutant construct ‘BNE’ (binding not editing) that allows ADAR binding but prevents editing (31) (Supplementary Figure S2). This construct was spliced as efficiently as the wild-type construct and was not affected by ADAR2 cotransfection. This demonstrates that an inosine at the R/G site but not binding of ADAR2 is responsible for reduced splicing of intron 13 (Figure 2B and C).
Editing at the R/G site prevents erroneous splicing
GluR-B is alternatively spliced downstream of the R/G site. Two splice conformations exist in vivo. The flip conformation fuses exon 13 to exons 15 and 16 while in the flop conformation exon 13 is fused to exons 14 and 16 (34). Conceptually, it is possible that editing in exon 13 preferentially suppresses splicing of the adjacent intron 13 but leaves splicing of intron 14 unaffected. If this was the case, editing might lead to preferential exclusion of exon 14 and inclusion of exon 15 (flip conformation). Therefore, to test whether R/G site editing influences alternative splicing, fragments spanning exons 13–16, resembling the edited, an uneditable, or wild-type state were introduced into the RFP-GFP vector and transfected into HEK293 cells. Using primers located in the RFP and GFP regions of the construct, two bands were detected by RT-PCR (Figure 3A). Sequencing revealed that the upper band contained correctly spliced fragments in both, the flip and flop conformation at approximately the same frequency (exons 14 and 15 are of identical size). The faster migrating band represented an erroneously spliced product, fusing exon 13 directly to exon 16, leading to a premature stop codon. Interestingly, the erroneously spliced product was most prominent in wild-type and the uneditable R/G constructs, while the ‘pre-edited’ fragment was predominantly spliced correctly. Taken together with the finding that editing of the R/G site decreases the efficiency of splicing, our observation suggests that a decrease in the speed of splicing facilitates alternative splice site choice, at least in the context of the reporter construct.
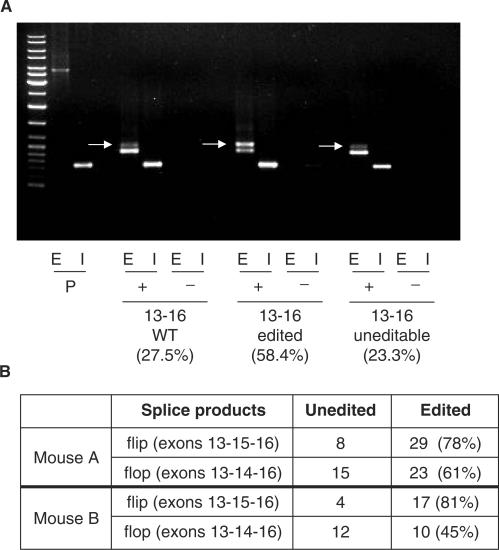
Figure 3.
Erroneous splicing and editing. (A) RT-PCR analysis of HeLa cells transfected with constructs expressing exons 13–16 with the R/G site in wild-type, ‘pre-edited’, and uneditable conformation. Exon (E) and intron (I) containing primers were used. Properly spliced products in both the flip and flop conformation run at equal height and are labeled by an arrow. An erroneously spliced product (exons 13–16/lower band) is most abundant in cells transfected with the wild-type and uneditable R/G constructs. The ‘pre-edited’ fragment was mainly spliced correctly. Constructs are indicated below the respective lanes. P: plasmid; +: with RTase; −: without RTase. The percentage of properly spliced product to total spliced product is indicated underneath each PCR reaction. (B) GluR-B cDNAs were amplified from two different mouse brains, cloned and sequenced individually. No clear correlation was observed between editing and alternative exon choice in mouse A. In mouse B, in contrast, a correlation between editing and splicing in the flip conformation can be observed. Given this variability amongst different mice, it seems that RNA editing and alternative exon choice are functionally unrelated.
The R/G site is located two nucleotides upstream of the 5′ splice-site of intron 13. Editing could therefore impair the activity of this splice donor site and selectively slow down splicing of intron 13 thus promoting preferential inclusion of exon 15, the flip conformation. To test for a possible correlation between R/G site editing and alternative splicing in vivo, cDNAs from adult mouse brain were amplified using primers located in exons 13 and 16, respectively. Here, considerable variation was observed between brain cDNAs from different mice. While a positive correlation between editing and splicing in the flip conformation was observed in the cDNA of one mouse (mouse B in Figure 3B), this correlation was rather weak in the cDNA obtained from another mouse (mouse A in Figure 3B). This indicates that both splicing and editing vary amongst different individuals and neuronal tissues. Most importantly, these two post-transcriptional events lack an obvious functional correlation as previously suggested (21).
Editing and splicing at the Q/R site
In contrast to the R/G site, lack of editing at the Q/R site selectively inhibits splicing of intron 11, and leads to nuclear accumulation of GluR-B pre-mRNA (23). To investigate the influence of editing at the Q/R site on splicing of intron 11, Q/R-containing genomic fragments spanning exons 11 and 12 (‘wild type’) were cloned into the RFP-GFP vector. Despite the use of different cells lines (HEK293, HeLa, neuroblastoma) splicing was only detected in a few cells, using the cell-based fluorescence splice assay (data not shown). Introduction of a guanosine at the Q/R site or removal of the ECS did not increase splicing efficiency. However, removal of large parts of intron 11 resulted in an increase in splicing activity (data not shown). The region removed also included an intronic editing hotspot 2, a target of ADAR2. Therefore, a construct was made where both the Q/R site and hotspot 2 in intron 11 were ‘pre-edited’. Observation of red and green fluorescence under the microscope indicated efficient splicing of this reporter. Unfortunately, a golgi-like localization pattern of all Q/R constructs precluded their quantitative analysis by FACS. Thus, to quantify splicing efficiencies, RT-PCR experiments were performed using intronic and exonic primer combinations. To remain in the exponential phase of amplification, 25 PCR cycles of different cDNA dilutions were run (Figure 4C). After electrophoresis (Figure 4A), band intensities were measured, and splicing efficiencies calculated (Figure 4B).
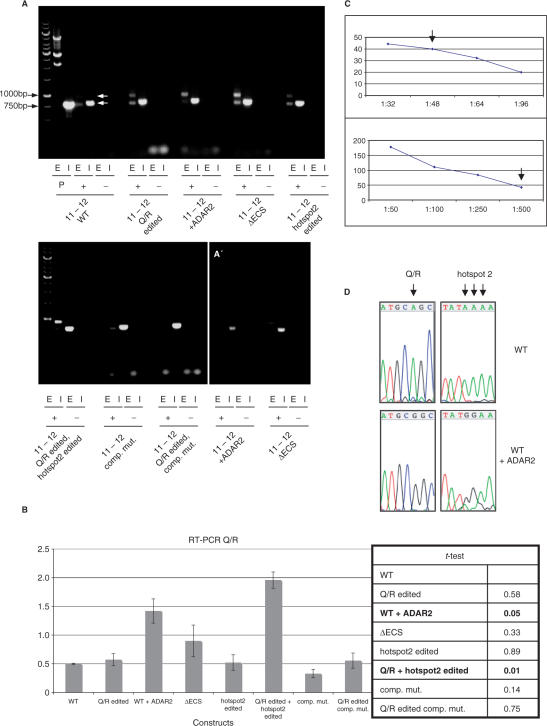
Figure 4.
Analysis of splice efficiencies of wt and mutated Q/R constructs.(A). RT-PCR of Q/R constructs transfected into HEK293 cells using 25 cycles, in the presence (+) or absence (−) of reverse transcriptase. The strongest signal is visible when the Q/R site and the intronic editing hotspot are edited concomitantly. The constructs are indicated below the respective lanes. E: exonic primer pair, spliced product. I: intronic primer pair, unspliced product. Arrows point to the spliced (upper arrow) and unspliced (lower arrow) PCR product, respectively. (A) Shorter exposure of two selected constructs, demonstrating that the PCR reactions with intronic primers have not yet reached stationary phase. (B) Calculated splicing efficiencies of three RT-PCR analysis are plotted in a graph. Band intensities were normalized for PCR product size and the relative splicing efficiencies were calculated as ratios of the splice product versus spliced plus unspliced. Note that the diminutive value for splicing efficiencies is due to the strong intronic signal. Co-transfection of wt Q/R with ADAR2, or a construct that is ‘pre-edited’ at both the Q/R site and the hotspot 2 spliced best. Editing at the Q/R site alone, deletion of the ECS, or a compensatory mutation only moderately altered splicing efficiency. Student's t-test indicates that the increase in splice efficiency upon cotranfection of ADAR2 or pre-editing of the Q/R site and hotspot 2 is significant. (C) Quantified band intensities of RT-PCRs with different DNA dilutions are plotted in the graphs. The linear relationship between the different DNA dilutions are shown for each primer pair. Arrows indicate the DNA dilution which was used for the RT-PCR in (A). Differing DNA dilutions for the exonic and intronic primer pairs were considered in the calculations for splicing efficiencies. (D) Co-transfection of ADAR2 results in editing of GluR-B at the Q/R site and the hotspot 2. The editing sites are indicated by arrows.
Wild-type and constructs ‘pre-edited’ at the Q/R site showed only limited splicing. Cotransfection of the wild-type construct with ADAR2 clearly increased splicing efficiencies at a statistically significant level. Removal of the ECS also led to a moderate increase in splicing which was found to be statistically insignificant. A construct ‘pre-edited’ at the intronic hotspot 2 alone showed no increase in splicing, while concomitant ‘pre-editing’ at intronic hotspot 2 and at the Q/R site showed the highest splicing efficiency that was also statistically significant (Figure 4A and B). The intronic editing site at hotspot 2 (AAA) is predicted to form a double-stranded structure by base pairing with a complementary region located at the intronic editing site at hotspot 1 (UUU) (Supplementary Figure S3). Editing of one of the two strands might help to open the double-stranded structure of this region, enabling access of factors that facilitate splicing. Therefore, a compensatory mutation was introduced around position 60, changing the three consecutive U residues opposing the editing hotspot to AAA, thus preventing base pairing with the adenosines to be edited. However, this mutant (compensatory mutation; Figure 4) displayed weak splicing even when combined with a “pre-edited” Q/R site (Q/R edited, compensatory mutation; Figure 4). This suggests that changes in the primary sequence rather than an alteration of the secondary structure are responsible for the observed increase in splicing efficiency upon editing of hotspot 2.
Sequencing of the spliced product derived from the wild-type fragment cotransfected with ADAR2 showed that exon 11 is correctly joined to exon 12 and that the Q/R site is edited. Sequencing of the unspliced precursor showed that hotspot 2 is efficiently edited upon cotransfection of ADAR2 (Figure 4D).
In addition to the PCR product of expected size, another, lower molecular weight band was observed in most RT-PCR reactions. Sequencing of this fragment revealed an erroneously spliced product. A cryptic splice site in exon 11 was used instead of the proper 5′ splice site in intron 11. The same erroneous fragment was isolated from mouse brain cDNA, which indicates that this splicing error can also occur in vivo (data not shown). Whether this error is due to under editing of GluR-B RNA cannot be proven, since the editing sites are spliced out in these RNAs.
Taken together, our data indicate that editing the Q/R base and an intronic hotspot is necessary and sufficient for efficient splicing. Conceivably, editing at hotspot 2 neutralizes a splice-repressor site.
DISCUSSION
Several findings suggest a tight co-transcriptional coupling of splicing and editing (22, 34–39).
Here, we show that RNA editing within the pre-mRNA encoding the B subunit of the mouse glutamate receptor can have dual effects on splicing. While editing at the Q/R site and at an adjacent intronic editing hotspot is a prerequisite for splicing of intron 11, editing at the R/G site downregulates splicing, possibly facilitating alternative splicing.
Splicing is suppressed by inosine at the R/G site
Suppression of splicing at the R/G site by ADAR2 has previously been observed in vitro, but not in vivo (26). Interestingly, the inhibitory effect of ADAR2 on in vitro splicing reactions was reduced by the addition of RNA-helicase A, suggesting that binding of ADAR2 can limit the accessibility of adjacent splice sites (26). In our assay, a clear reduction of splicing upon editing was also observed in vivo. Moreover, we demonstrate that the modified nucleotide but not ADAR2 binding down-regulates splicing (Figure 2). Several facts might account for the discrepancy of these two studies. First, our assay most likely allows a more sensitive readout, since even minor variations in splicing efficiency can be monitored in the large pool of cells analyzed by FACS. Moreover, in our study R/G site editing and splicing were analyzed in the context of naturally occurring flanking sequences rather than in the context of an adenoviral splice acceptor.
Interestingly, upon cotransfection of ADAR2 the extent of editing at the R/G site measured by comparing peak heights of RT-PCR sequencing reaction did not directly correlate with the observed decrease in splicing (compare Figure 2B and D). However, peak heights in sequencing reactions often vary depending on the sequence context and may thus not accurately reflect the abundance of a given base at a particular position.
The reduced splicing efficiency in the presence of an edited R/G site might be explained by the fact that the editing site is only one nucleotide away from the 5′ splice site of intron 13. Editing could possibly interfere with the base pairing of U1 snRNA. In fact, the wild-type sequence resembles the consensus sequence for 5′ splice sites more closely than its edited counterpart (40). However, the edited R/G site and surrounding sequences also resemble a splice silencer sequence to which hnRNP A1 can bind (41). (Splice silencer consensus: GGCAGGGUGG; edited R/G site and adjacent 5′ splice site: UUAGGGUGG). It therefore also seems possible that editing introduces a binding site for a splice silencer which mediates the observed effect on splicing.
Editing at the R/G facilitates alternative splicing
In the reporter assay, erroneously spliced RNAs skipping both alternatively spliced exons were observed in the absence of editing. Editing-induced reduction in the speed of splicing of intron 13 might facilitate coordinated activation of only one of the branch points located in either intron 13 or 14, consequently allowing alternative splicing to occur (Figure 5). This notion is consistent with the recent finding that editing but not splicing at the R/G site is regulated by the CTD of RNA-polymerase II. It thus seems that deposition of ADAR2 is regulated cotranscriptionally by RNA-polymerase II to facilitate alternative splicing of the downstream exons (27). Interestingly, no clear correlation between exon choice and editing could be observed. Investigation of several mouse brain cDNA samples showed a high variability amongst different samples. While some samples showed a clear correlation between editing and splicing in the flip conformation, this correlation was lacking from other samples. It thus appears that editing and splice site choice are independent events at the R/G site.
Figure 5.
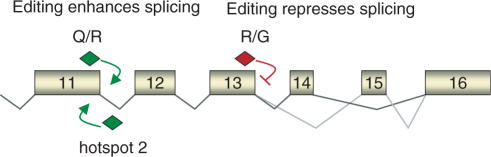
Regulation of splicing by editing in GluR-B pre-mRNA. At the Q/R site, editing of the Q/R base and an intronic hotspot is required for efficient splicing. A double-stranded structure is potentially resolved at the Q/R site, while a splice repressor is inactivated at the intronic editing hotspot 2. The edited nucleotides are sufficient to stimulate splicing. Editing at the R/G site represses splicing and enhances proper alternative splicing. An edited nucleotide at the R/G site is sufficient to suppress splicing. ADAR binding is not required.
However, editing can regulate alternative splicing such as in the serotonin 2C receptor pre-mRNA (42,28). A strong correlation between editing efficiency and downstream alternative splice site choice has also been observed in Drosophila (29).
An intronic editing hotspot is required for Q/R-site splicing
Lack of ADAR2 inhibits splicing of the intron downstream of the GluR-B Q/R site (23). Here, we have shown that editing of the intronic hotspot 2 located at position +262/263/264 is crucial for splicing of the surrounding intron. The importance of this region can also be deduced from the high sequence conservation from Tetraodon to man.
Previously, it was assumed that editing of the Q/R site itself was the only requirement for successful splicing of intron 11, since a ‘pre-edited’ GluR-B allele introduced into ADAR2 knockout mice shows normal splicing (23,43,44). However, in this allele the intronic editing hotspot 2 was deleted by a neo cassette (44). Both editing hotspots 1 (+60) and 2 (+262/263/264) are located within imperfect inverted repeats located upstream of the ECS (Supplementaary Figure S3) (22,45). Base pairing of these inverted repeats has been shown to be essential for Q/R editing possibly by aiding the base pairing of the ECS and the Q/R site. ECS mutants, in turn, display a reduced editing efficiency at the intronic editing hotspots assayed in tissue culture cells (22). Editing of the intronic hot-spot at +262 might serve as a molecular switch: only after successful base pairing of all complementary sequences the Q/R site and the intronic site can be edited, which in turn would allow splicing to proceed. This control mechanism could explain the high editing levels at the Q/R site observed in vivo, since only messages that have been modified by ADAR2 are spliced and exported to the cytoplasm (Figure 5).
The mechanism by which editing of intronic hotspot 2 regulates splicing is not clear at this point. Alterations in the secondary structure in intron 11 alone are not sufficient to promote splicing. Instead, editing at the intronic hotspot 2 might serve to modulate a splice silencer sequence. As for the R/G site, the base exchange by itself is sufficient to enhance splicing, indicating that binding of ADAR2 is not required for efficient splicing.
It is obvious that RNA editing plays a crucial role in generating protein diversity. Our study demonstrates that this is not only achieved by amino acid exchanges through editing in coding sequences, but also by regulating splicing efficiency and alternative splicing decisions.
SUPPLEMENTARY DATA
Supplementory Data are avialable at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Teresa Spreitzer for excellent technical assistance and Martin Xaver for help with the quantification of PCR bands. Rat ADAR2 and mouse ADAR1 expression plasmids were kindly provided by Ron Emeson and Mary O’Connell, respectively. This work was supported by the Austrian Science Foundation grant no. SFB 1706. Funding to pay this Open Access publication changes for this article was provided by the Austrian Science Foundation.
Conflict of interest statment. None declared.
REFERENCES
- 1.Chervitz SA, Aravind L, Sherlock G, Ball CA, Koonin EV, Dwight SS, Harris MA, Dolinski K, Mohr S, et al. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 3.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner RW, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan NF, Khalili K, Kim SU, Perussia B, et al. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol. Cell. Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connell MA, Keegan LP. Drosha versus ADAR: wrangling over pri-miRNA. Nat. Struct. Mol. Biol. 2006;13:3–4. doi: 10.1038/nsmb0106-3. [DOI] [PubMed] [Google Scholar]
- 8.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 9.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 10.Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA [letter] RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 11.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O'Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 13.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 16.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 17.Hume R, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 18.Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 19.Greger IH, Khatri L, Kong X, Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- 20.Koehler M, Kornau HC, Seeburg PH. The organization of the gene for the functionally dominant alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor subunit GluR-B. J. Biol. Chem. 1994;269:17367–17370. [PubMed] [Google Scholar]
- 21.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 25.Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 26.Bratt E, Ohman M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl Acad. Sci. USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal R, Stormo GD. Editing efficiency of a Drosophila gene correlates with a distant splice site selection. RNA. 2005;11:563–566. doi: 10.1261/rna.7280605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 31.Kallman AM, Sahlin M, Ohman M. ADAR2 A—>I editing: site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003;31:4874–4881. doi: 10.1093/nar/gkg681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strehblow A, Hallegger M, Jantsch MF. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol. Cell. 2002;13:3822–3835. doi: 10.1091/mbc.E02-03-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 34.Seeburg PH, Higuchi M, Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Brain Res. Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 35.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 36.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 37.Reenan RA, Hanrahan CJ, Barry G. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 38.Eckmann CR, Jantsch MF. The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J. Cell. Biol. 1999;144:603–615. doi: 10.1083/jcb.144.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc. Natl Acad. Sci. USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairbrother WG, Chasin LA. Human genomic sequences that inhibit splicing. Mol. Cell Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flomen R, Knight J, Sham P, Kerwin R, Makoff A. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nuleic Acids Res. 2004;32:2113–2122. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing- deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 44.Kask K, Zamanillo D, Rozov A, Burnashev N, Sprengel R, Seeburg PH. The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc. Natl Acad. Sci. USA. 1998;95:13777–13782. doi: 10.1073/pnas.95.23.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J-H, Sklar P, Axel R, Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995;374:77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]