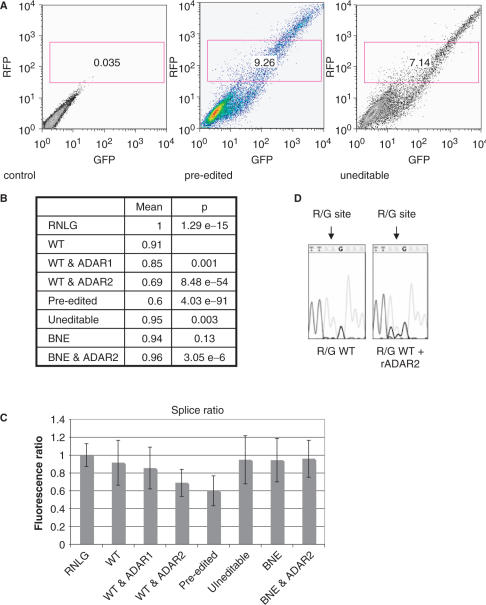
Figure 2.
FACS analysis of R/G constructs transfected into HEK293 cells. (A) 2D plot of untransfected cells (control), cells transfected with a vector containing exons 13 through 14 including the R/G site in a pre edited, and uneditable state. Constitutive red fluorescence is plotted along the y-axis while green fluorescence (only visible after successful splicing) is plotted along the x-axis. Only cells expressing solid RFP expression were chosen and gated for further analysis. (B) Red to green fluorescence ratios of individual cells in the gated window shown in (A) were calculated. The mean of these ratios is given. For clarity, the mean fluorescence ratio of cells transfected with ‘empty vector’ (RNLG) was set to 1. Wild-type and mutant R/G constructs spanning exons 13 through 14, and plasmids expressing ADAR1 or ADAR2 are shown. A ‘pre-edited’ R/G construct and a wild-type construct cotransfected with ADAR2 are weakly spliced. However, a construct where ADAR can bind but is unable to edit (binding but no editing +ADAR2) showed GFP fluorescence comparable to the wild-type construct, indicating that an inosine at the R/G site and not ADAR binding is responsible for splice suppression. Student's t-test P-values indicate that populations of transfected cells differed significantly from cells transfected with the wild-type construct with the exception of cells transfected with the construct that could be bound but not edited by ADAR2 (BNE). (C) Bar diagram and SD of RFP:GFP fluorescence ratios shown in (B). (D) Cotransfection of ADAR2 results in editing of GluR-B at the R/G site. Sequencing of RT-PCR products revealed that wild-type R/G fragments display no G peak at the R/G site whereas cotransfection of ADAR 2 results in editing. The R/G site is marked by an arrow.